"crystallin, beta B1, like 3"
ZFIN 
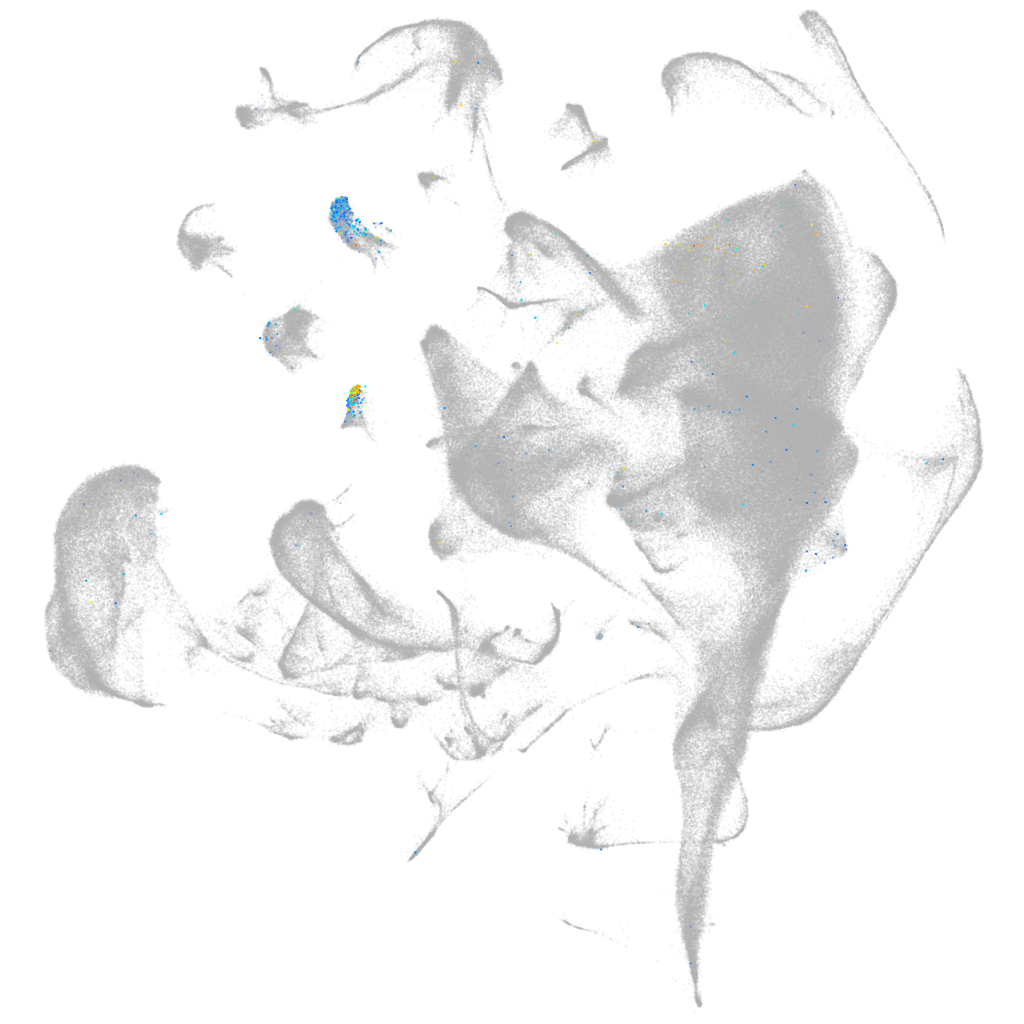
Other cell groups


all cells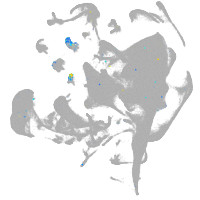 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 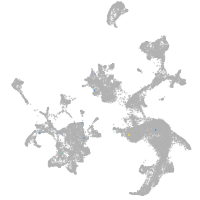 |
pronephros  |
neural |
spinal cord / glia |
eye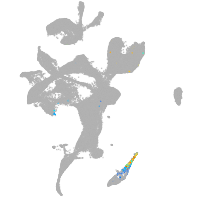 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cryba1l2 | 0.390 | hmgb1b | -0.041 |
| mipa | 0.363 | h2afva | -0.037 |
| lim2.4 | 0.362 | hmgb3a | -0.037 |
| lim2.5 | 0.346 | marcksb | -0.037 |
| cx23 | 0.342 | nucks1a | -0.037 |
| crybb1l1 | 0.339 | si:ch211-288g17.3 | -0.036 |
| capn3a | 0.334 | hp1bp3 | -0.035 |
| lim2.3 | 0.333 | si:dkey-42i9.4 | -0.035 |
| crybb1l2 | 0.332 | hnrnpa0a | -0.034 |
| mipb | 0.331 | tuba1c | -0.034 |
| cryba2a | 0.328 | gnb1b | -0.033 |
| BX548032.3 | 0.322 | hdac1 | -0.033 |
| crygmx | 0.315 | jpt1b | -0.032 |
| cryba1b | 0.314 | si:ch1073-429i10.3.1 | -0.032 |
| cryba1l1 | 0.308 | nova2 | -0.031 |
| gpx9 | 0.304 | tmeff1b | -0.031 |
| cryba1a | 0.302 | tuba1a | -0.031 |
| crybb1 | 0.297 | anp32a | -0.030 |
| lim2.1 | 0.296 | atrx | -0.030 |
| crygn2 | 0.294 | chd4a | -0.030 |
| gja8b | 0.294 | fxyd6l | -0.030 |
| si:ch211-214j24.14 | 0.289 | rtn1a | -0.030 |
| gja8a | 0.286 | si:ch73-46j18.5 | -0.030 |
| cryba2b | 0.278 | srsf3b | -0.030 |
| grifin | 0.277 | CABZ01080702.1 | -0.029 |
| si:ch211-255g12.6 | 0.277 | cbx5 | -0.029 |
| cryba4 | 0.275 | gpm6aa | -0.029 |
| BFSP1 | 0.265 | ier5 | -0.029 |
| endou2 | 0.264 | pbx4 | -0.029 |
| crygm2d10 | 0.260 | seta | -0.029 |
| crygm2d13 | 0.260 | tubb5 | -0.029 |
| crygmxl2 | 0.254 | acin1a | -0.028 |
| bfsp2 | 0.246 | elavl3 | -0.028 |
| crygm2d8 | 0.245 | si:ch211-137a8.4 | -0.028 |
| crygm2d12 | 0.242 | si:ch211-51e12.7 | -0.028 |