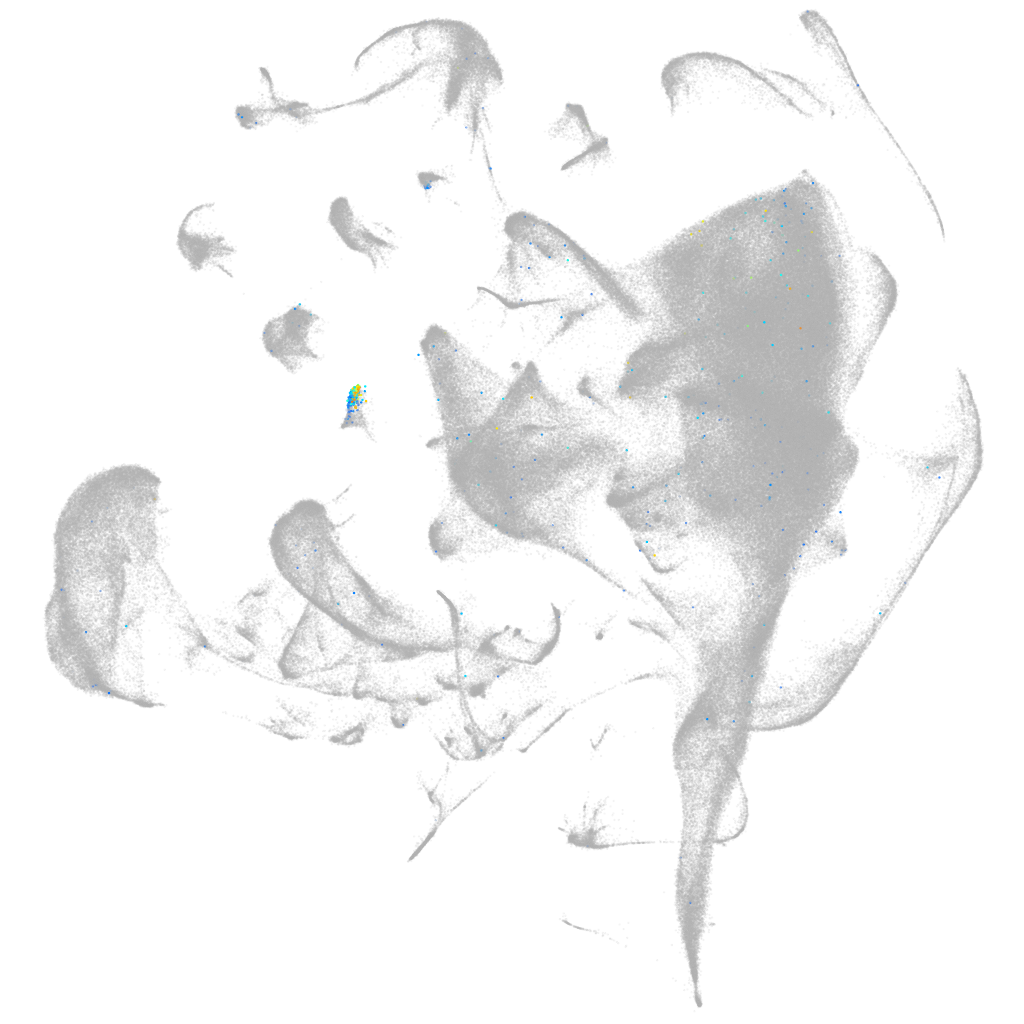
"crystallin, beta A1, like 2"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lim2.4 | 0.464 | h2afva | -0.025 |
| mipa | 0.459 | fabp3 | -0.023 |
| crybb1l1 | 0.448 | hmgb1b | -0.022 |
| crybb1l2 | 0.438 | si:ch211-288g17.3 | -0.022 |
| cryba2a | 0.431 | nucks1a | -0.021 |
| lim2.3 | 0.431 | si:dkey-42i9.4 | -0.021 |
| capn3a | 0.428 | anp32a | -0.020 |
| cx23 | 0.426 | ctsla | -0.020 |
| BX548032.3 | 0.423 | marcksb | -0.020 |
| lim2.5 | 0.414 | fxyd6l | -0.019 |
| cryba1b | 0.413 | ier5 | -0.019 |
| mipb | 0.412 | mcl1a | -0.019 |
| crygmx | 0.402 | adh5 | -0.018 |
| cryba1l1 | 0.400 | cbx5 | -0.018 |
| crybb1l3 | 0.390 | hp1bp3 | -0.018 |
| lim2.1 | 0.387 | stmn1a | -0.018 |
| crygn2 | 0.382 | aldob | -0.017 |
| gpx9 | 0.382 | CABZ01080702.1 | -0.017 |
| crybb1 | 0.379 | chd4a | -0.017 |
| gja8b | 0.371 | cx43.4 | -0.017 |
| crygmxl2 | 0.362 | hdac1 | -0.017 |
| cryba4 | 0.361 | hmgb3a | -0.017 |
| cryba2b | 0.360 | hnrnpa0a | -0.017 |
| crygm2d10 | 0.352 | rbbp4 | -0.017 |
| gja8a | 0.351 | ube2ib | -0.017 |
| grifin | 0.351 | atp5if1a | -0.016 |
| crygm2d13 | 0.351 | btg2 | -0.016 |
| endou2 | 0.347 | gabarapb | -0.016 |
| BFSP1 | 0.343 | ier2b | -0.016 |
| si:ch211-255g12.6 | 0.336 | nova2 | -0.016 |
| crygm2d8 | 0.329 | pcna | -0.016 |
| crygm2d12 | 0.327 | seta | -0.016 |
| BX545855.1 | 0.321 | si:ch211-137a8.4 | -0.016 |
| bfsp2 | 0.305 | si:ch73-46j18.5 | -0.016 |
| nags | 0.296 | si:dkey-56m19.5 | -0.016 |