N-acetylglutamate synthase
ZFIN 
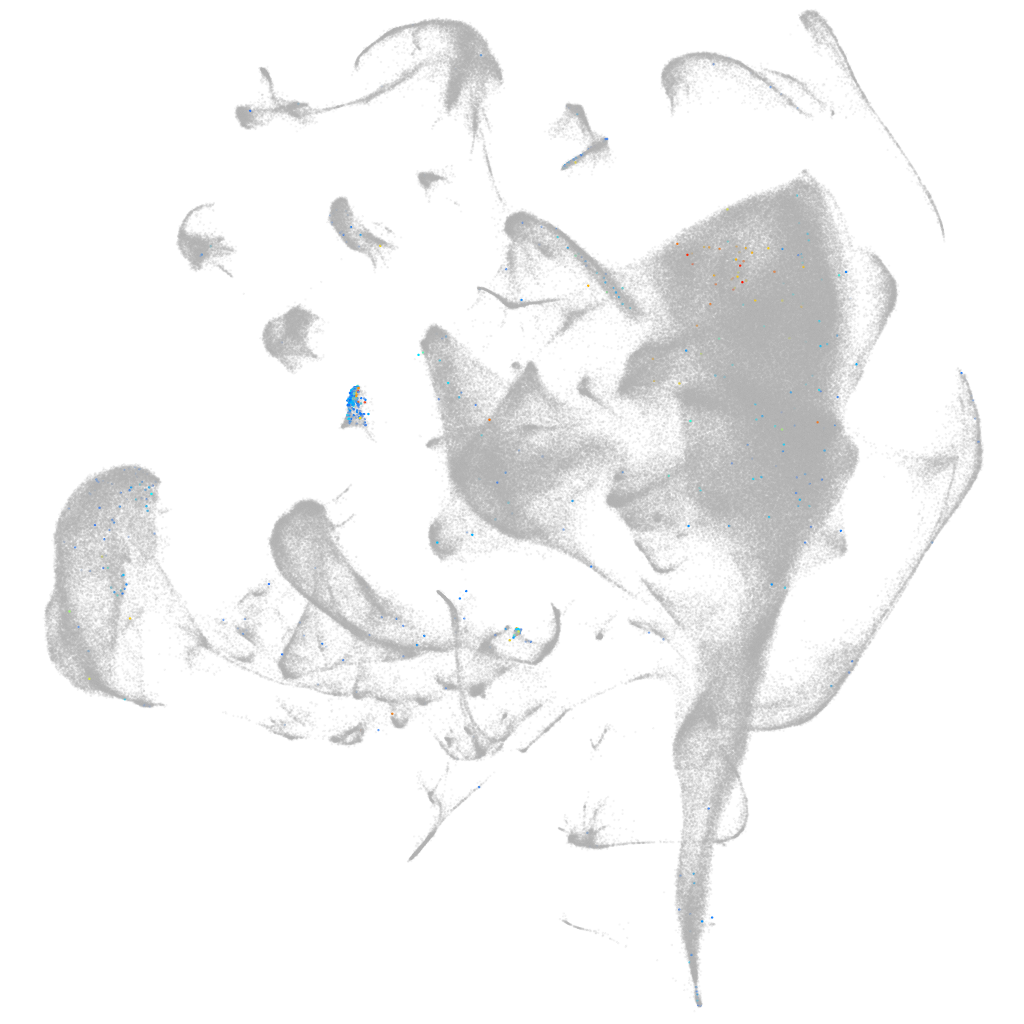
Other cell groups


all cells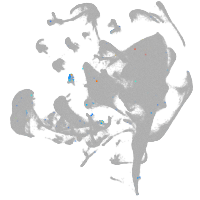 |
blastomeres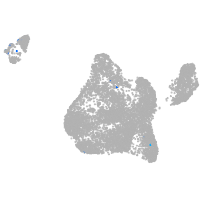 |
PGCs |
endoderm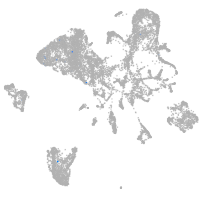 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye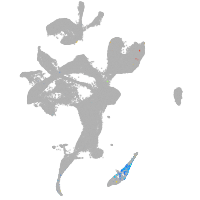 |
taste / olfactory |
otic / lateral line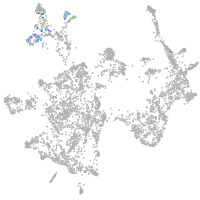 |
epidermis |
periderm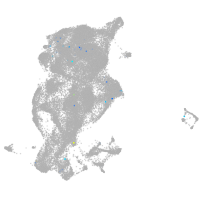 |
fin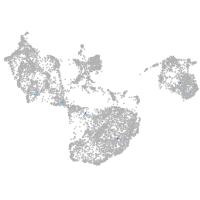 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gja8b | 0.308 | hmgb1b | -0.022 |
| cx23 | 0.299 | fabp3 | -0.020 |
| cryba1l2 | 0.296 | marcksb | -0.020 |
| gja8a | 0.289 | si:ch211-288g17.3 | -0.020 |
| lim2.4 | 0.282 | si:dkey-56m19.5 | -0.019 |
| cryba2a | 0.273 | tuba1c | -0.018 |
| lim2.1 | 0.267 | hmgb3a | -0.017 |
| crybb1l1 | 0.266 | mdkb | -0.017 |
| lim2.3 | 0.266 | nova2 | -0.017 |
| mipa | 0.263 | cd81a | -0.016 |
| cryba1l1 | 0.255 | elavl3 | -0.016 |
| cps1 | 0.254 | gpm6aa | -0.016 |
| crybb1l2 | 0.254 | h2afva | -0.016 |
| cryba1b | 0.249 | mdka | -0.016 |
| crybb1 | 0.247 | nucks1a | -0.016 |
| BFSP1 | 0.242 | rtn1a | -0.016 |
| BX548032.3 | 0.241 | si:ch211-137a8.4 | -0.016 |
| crybb1l3 | 0.238 | sox11a | -0.016 |
| gpx9 | 0.238 | tmeff1b | -0.016 |
| mipb | 0.235 | tubb5 | -0.016 |
| cryba4 | 0.228 | adh5 | -0.015 |
| crygmx | 0.227 | hp1bp3 | -0.015 |
| crygn2 | 0.220 | si:dkey-42i9.4 | -0.015 |
| prox2 | 0.220 | stmn1a | -0.015 |
| otc | 0.216 | tmsb | -0.015 |
| cryba2b | 0.211 | cx43.4 | -0.014 |
| gja3 | 0.210 | FO082781.1 | -0.014 |
| lim2.5 | 0.206 | mex3b | -0.014 |
| capn3a | 0.201 | pmp22a | -0.014 |
| endou2 | 0.193 | sox11b | -0.014 |
| crygmxl2 | 0.179 | stmn1b | -0.014 |
| CR356246.1 | 0.178 | tp53inp2 | -0.014 |
| grifin | 0.175 | tuba1a | -0.014 |
| nrl | 0.171 | mfap2 | -0.014 |
| bfsp2 | 0.169 | CABZ01080702.1 | -0.013 |