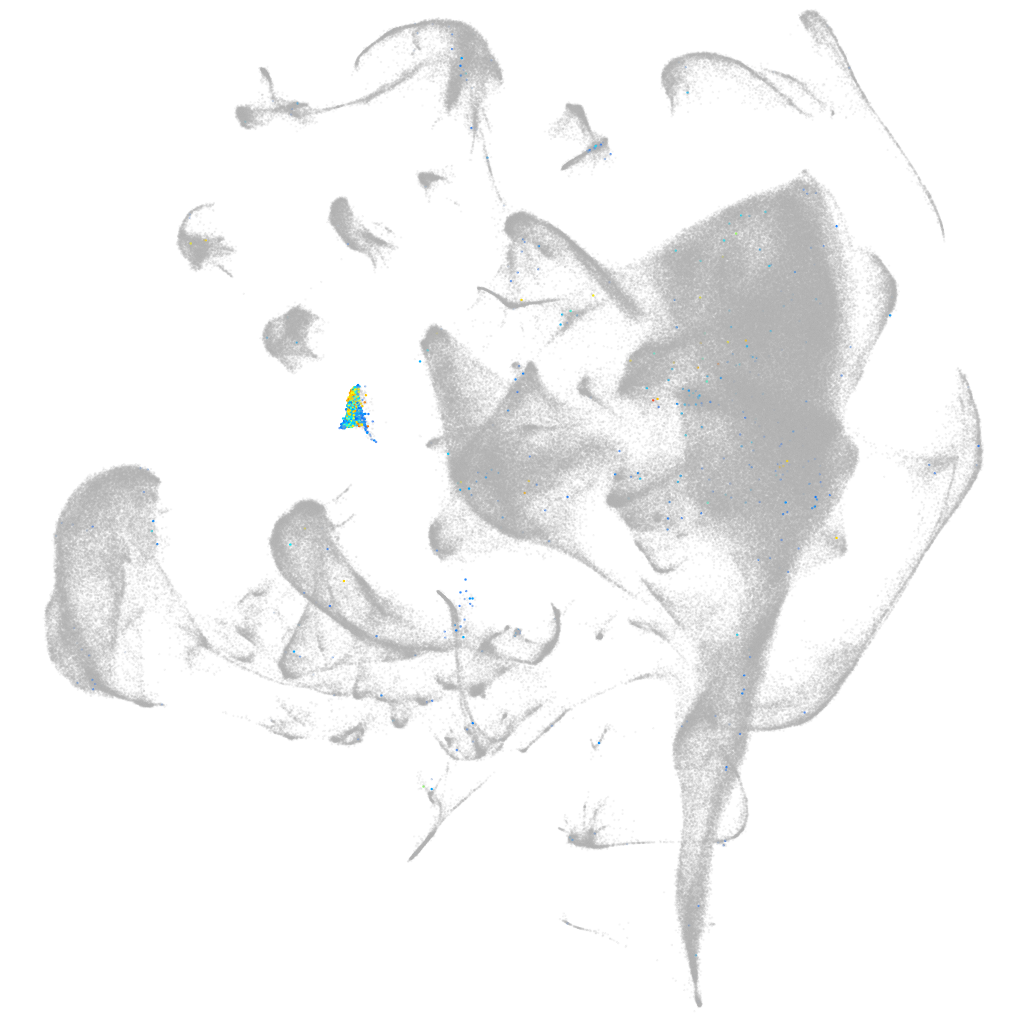
gap junction protein alpha 8 paralog b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cps1 | 0.509 | fabp3 | -0.054 |
| gja8a | 0.499 | mdka | -0.042 |
| si:ch211-219a15.3 | 0.486 | h2afva | -0.034 |
| crybb1 | 0.477 | sparc | -0.033 |
| CR356246.1 | 0.476 | pmp22a | -0.031 |
| cryba2a | 0.472 | zfp36l1a | -0.031 |
| LOC110438076 | 0.470 | adh5 | -0.030 |
| prox2 | 0.465 | tuba1a | -0.030 |
| col28a1b | 0.452 | atp5if1a | -0.029 |
| mipb | 0.451 | dbi | -0.029 |
| lim2.1 | 0.445 | meis1b | -0.029 |
| foxe3 | 0.443 | si:dkey-42i9.4 | -0.029 |
| cx23 | 0.440 | cd63 | -0.028 |
| cryba1l1 | 0.433 | fabp7a | -0.028 |
| BFSP1 | 0.423 | si:dkey-56m19.5 | -0.028 |
| sparcl1 | 0.423 | mfap2 | -0.028 |
| lim2.4 | 0.421 | jpt1b | -0.027 |
| cryba4 | 0.411 | mdkb | -0.027 |
| cryba1b | 0.409 | nova2 | -0.027 |
| lim2.3 | 0.401 | tuba1c | -0.027 |
| crybb1l1 | 0.399 | CABZ01080702.1 | -0.026 |
| FO681288.1 | 0.382 | marcksb | -0.026 |
| mipa | 0.382 | si:ch211-137a8.4 | -0.026 |
| crygmx | 0.378 | sox11a | -0.026 |
| crybb1l2 | 0.375 | ephb3 | -0.026 |
| slc7a11 | 0.375 | si:dkey-261h17.1 | -0.026 |
| cryba1l2 | 0.371 | hmgb1b | -0.025 |
| gja3 | 0.366 | mcl1a | -0.025 |
| otc | 0.363 | col5a1 | -0.025 |
| crygn2 | 0.341 | si:ch211-286o17.1 | -0.025 |
| cryba2b | 0.338 | agrn | -0.024 |
| nags | 0.308 | col1a2 | -0.024 |
| gpx9 | 0.299 | cx43.4 | -0.024 |
| crybb1l3 | 0.294 | krt18b | -0.024 |
| col4a3 | 0.287 | tmeff1b | -0.024 |