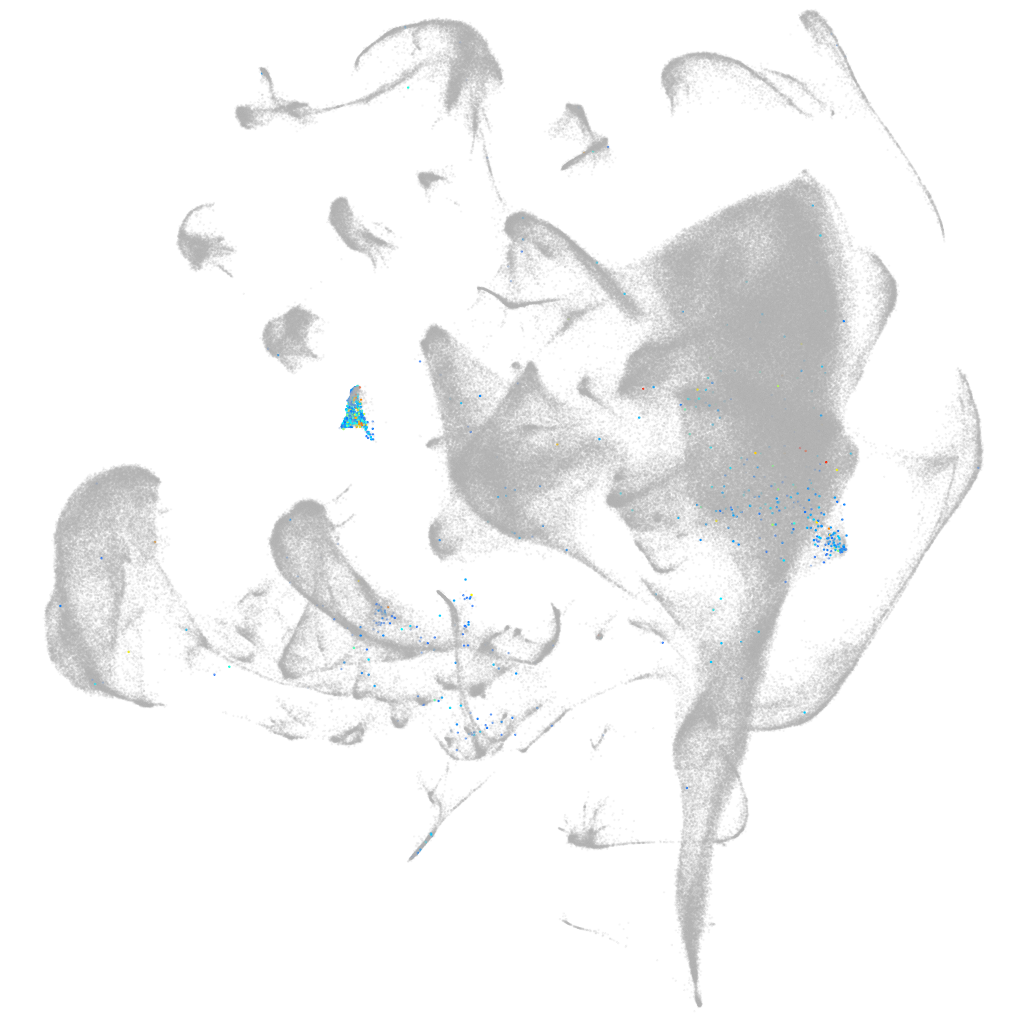
Other cell groups


all cells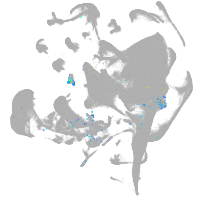 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye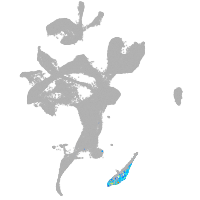 |
taste / olfactory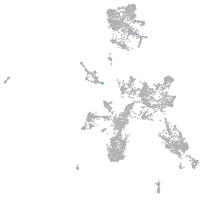 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gja8b | 0.470 | fabp3 | -0.030 |
| foxe3 | 0.415 | fabp7a | -0.027 |
| CR356246.1 | 0.369 | mdka | -0.027 |
| cps1 | 0.365 | sparc | -0.025 |
| col28a1b | 0.349 | krt18b | -0.022 |
| lim2.1 | 0.349 | si:dkey-56m19.5 | -0.022 |
| prox2 | 0.320 | tuba1a | -0.022 |
| crybb1 | 0.302 | tuba8l3 | -0.022 |
| FO681288.1 | 0.302 | col1a2 | -0.021 |
| si:ch211-219a15.3 | 0.301 | pmp22a | -0.021 |
| slc7a11 | 0.285 | col5a1 | -0.021 |
| BFSP1 | 0.277 | mfap2 | -0.021 |
| cx23 | 0.275 | cd63 | -0.020 |
| sparcl1 | 0.256 | col1a1a | -0.020 |
| gja8a | 0.247 | cpn1 | -0.020 |
| cryba4 | 0.244 | jpt1b | -0.020 |
| cryba1l1 | 0.242 | lrrc17 | -0.020 |
| cryba2a | 0.228 | tmem119b | -0.020 |
| pitx3 | 0.226 | col1a1b | -0.019 |
| otc | 0.215 | mdkb | -0.019 |
| gja3 | 0.208 | tmsb | -0.019 |
| col28a1a | 0.200 | twist1a | -0.019 |
| cryba2b | 0.197 | bhmt | -0.019 |
| lim2.3 | 0.190 | adh5 | -0.018 |
| cryba1b | 0.188 | ckbb | -0.018 |
| wnt2ba | 0.185 | col5a2a | -0.018 |
| col4a3 | 0.184 | nova2 | -0.018 |
| col4a4 | 0.183 | vwde | -0.018 |
| lim2.4 | 0.176 | ephb3 | -0.018 |
| crybb1l1 | 0.170 | marcksl1a | -0.018 |
| si:ch211-59p23.1 | 0.168 | si:dkey-261h17.1 | -0.018 |
| fabp11a | 0.166 | CU467822.1 | -0.017 |
| mipb | 0.164 | ier2b | -0.017 |
| hmx4 | 0.160 | mcl1a | -0.017 |
| nags | 0.159 | prrx1a | -0.017 |