"crystallin, beta A1a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis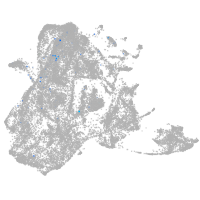 |
periderm |
fin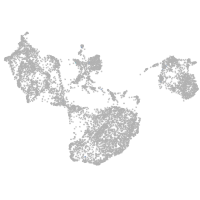 |
ionocytes / mucous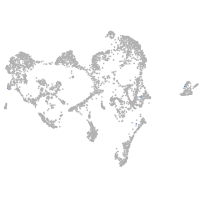 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crybb1l3 | 0.302 | marcksb | -0.030 |
| cryba1l2 | 0.260 | nucks1a | -0.030 |
| capn3a | 0.253 | hmgb1b | -0.027 |
| lim2.5 | 0.249 | si:ch211-288g17.3 | -0.026 |
| BX548032.3 | 0.246 | hdac1 | -0.025 |
| mipa | 0.223 | hnrnpa0a | -0.025 |
| lim2.4 | 0.217 | acin1a | -0.024 |
| crybb1l1 | 0.205 | hmgb3a | -0.024 |
| lim2.3 | 0.205 | hp1bp3 | -0.024 |
| gpx9 | 0.204 | chd4a | -0.023 |
| cx23 | 0.203 | nova2 | -0.023 |
| crygm2d10 | 0.203 | cx43.4 | -0.022 |
| crybb1l2 | 0.202 | h2afva | -0.022 |
| mipb | 0.200 | hnrnpa1a | -0.022 |
| grifin | 0.199 | tuba1a | -0.022 |
| si:ch211-255g12.6 | 0.199 | tuba1c | -0.022 |
| crygm2d13 | 0.198 | zc4h2 | -0.022 |
| crygm2d12 | 0.195 | anp32a | -0.021 |
| crygmx | 0.195 | atrx | -0.021 |
| cryba2a | 0.193 | cbx3a | -0.021 |
| crygm2d8 | 0.189 | ewsr1a | -0.021 |
| endou2 | 0.189 | gnb1b | -0.021 |
| cryba1b | 0.188 | seta | -0.021 |
| crygmxl2 | 0.188 | si:ch211-51e12.7 | -0.021 |
| lim2.1 | 0.188 | snrpf | -0.021 |
| BX545855.1 | 0.183 | srsf3b | -0.021 |
| cryba1l1 | 0.181 | tmeff1b | -0.021 |
| crygm2d5 | 0.177 | tubb5 | -0.021 |
| crygn2 | 0.176 | hnrnpub | -0.020 |
| bfsp2 | 0.175 | jpt1b | -0.020 |
| crybb1 | 0.175 | nono | -0.020 |
| crygm2d21 | 0.174 | pbx4 | -0.020 |
| crygm2d7 | 0.174 | rtn1a | -0.020 |
| BFSP1 | 0.169 | si:ch73-46j18.5 | -0.020 |
| crygm2d18 | 0.167 | smarce1 | -0.020 |




