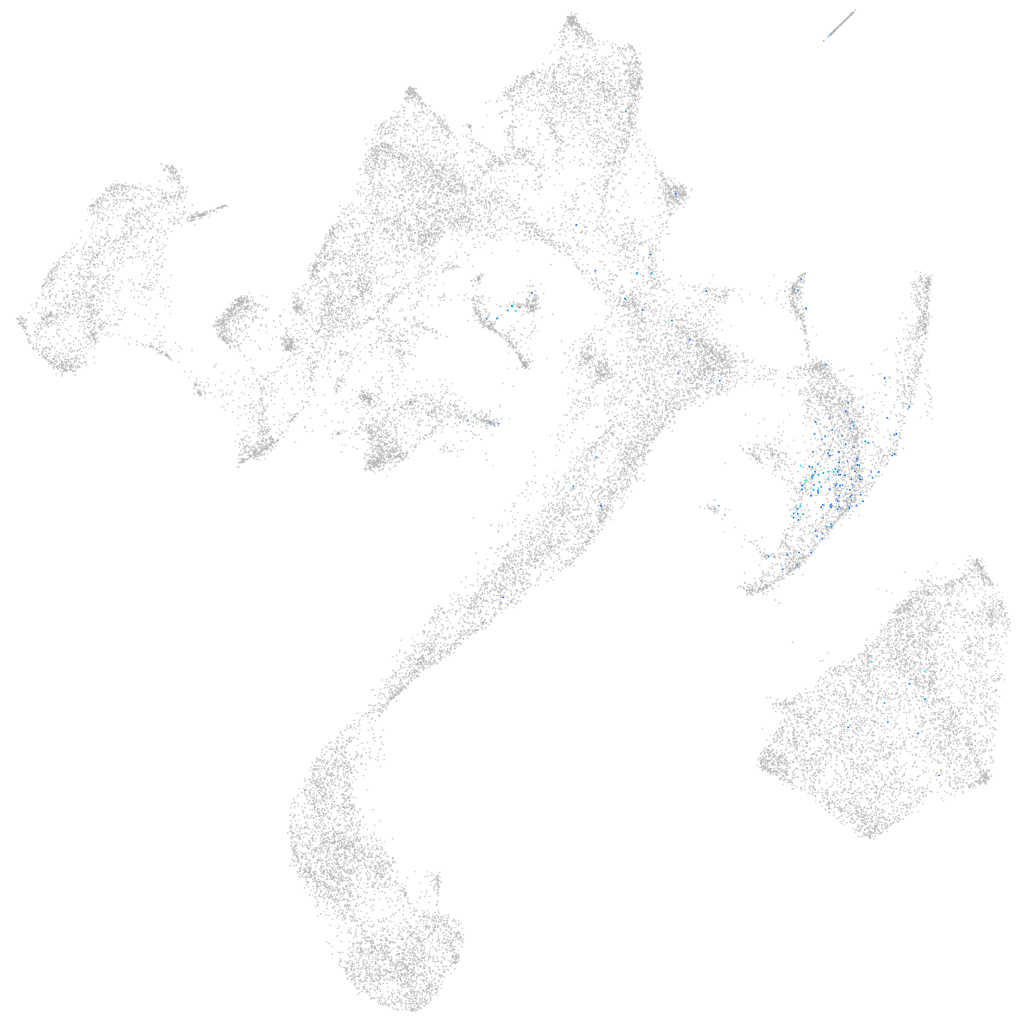
"Pim proto-oncogene, serine/threonine kinase, related 201"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| her1 | 0.180 | fabp3 | -0.056 |
| itm2cb | 0.161 | actc1b | -0.054 |
| her12 | 0.161 | ttn.2 | -0.050 |
| hoxd10a | 0.160 | eef1da | -0.049 |
| her7 | 0.154 | pabpc4 | -0.047 |
| tbx16l | 0.154 | ckma | -0.046 |
| msgn1 | 0.148 | ak1 | -0.046 |
| tnfrsf9a | 0.144 | ckmb | -0.045 |
| fzd10 | 0.140 | atp2a1 | -0.045 |
| si:dkeyp-46h3.3 | 0.137 | aldoab | -0.045 |
| hoxb10a | 0.136 | bhmt | -0.045 |
| pcdh8 | 0.133 | tmem38a | -0.044 |
| ephb3 | 0.119 | atp5l | -0.044 |
| nid2a | 0.117 | ttn.1 | -0.043 |
| hoxd11a | 0.116 | sparc | -0.043 |
| hoxa11a | 0.113 | tnnc2 | -0.043 |
| htr5aa | 0.112 | gapdh | -0.043 |
| greb1 | 0.110 | klhl41b | -0.042 |
| LO016987.2 | 0.110 | si:dkey-16p21.8 | -0.042 |
| hes6 | 0.110 | desma | -0.042 |
| trim35-25 | 0.107 | fxr2 | -0.041 |
| LOC108190033 | 0.106 | eno1a | -0.041 |
| LOC108179710 | 0.105 | srl | -0.041 |
| hoxd12a | 0.105 | cox17 | -0.041 |
| phkg2 | 0.105 | mylpfa | -0.041 |
| apoc1 | 0.104 | gamt | -0.041 |
| dlc | 0.103 | atp5if1b | -0.041 |
| cyp26a1 | 0.102 | rplp2 | -0.041 |
| fn1b | 0.100 | COX3 | -0.041 |
| BX005254.3 | 0.100 | tpma | -0.041 |
| BX649282.3 | 0.099 | acta1b | -0.041 |
| vox | 0.099 | gatm | -0.040 |
| crp7 | 0.097 | cox7c | -0.040 |
| fbln1 | 0.096 | CABZ01078594.1 | -0.040 |
| fn1a | 0.096 | vwde | -0.040 |