neurotrophin receptor associated death domain
ZFIN 
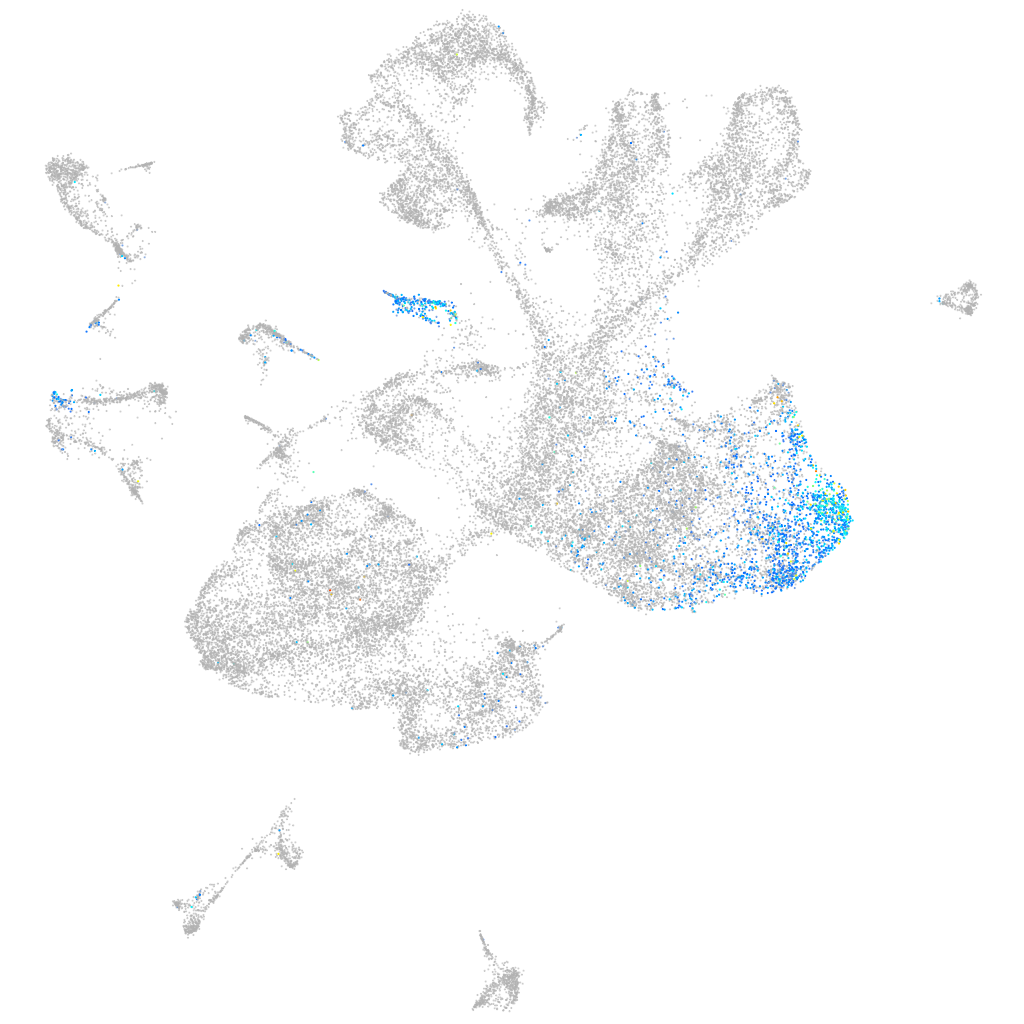
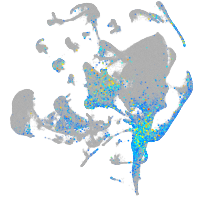
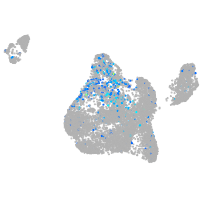
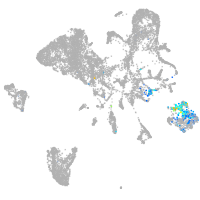
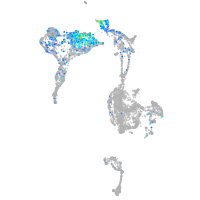
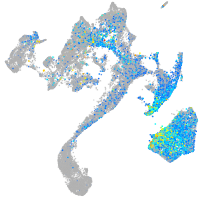
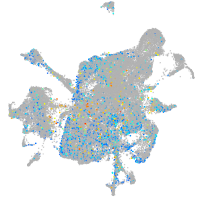
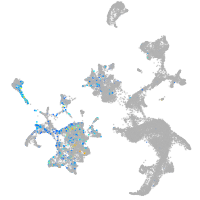
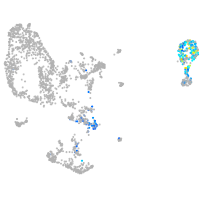
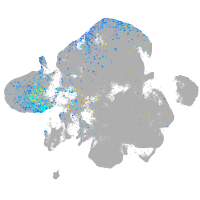
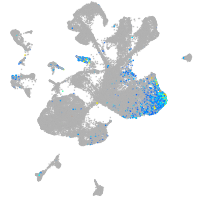
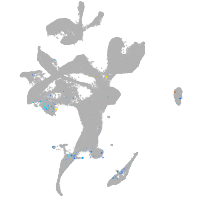
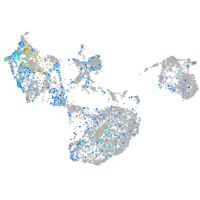
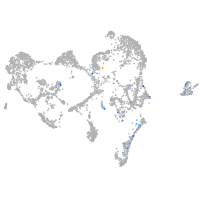
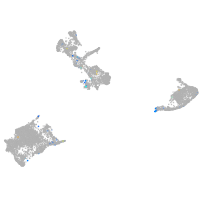
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdx4 | 0.549 | gpm6aa | -0.227 |
| sp5l | 0.468 | ckbb | -0.200 |
| hspb1 | 0.467 | CR383676.1 | -0.182 |
| apela | 0.440 | rtn1a | -0.179 |
| chrd | 0.396 | marcksl1b | -0.176 |
| BX927258.1 | 0.392 | fabp7a | -0.169 |
| fgf8a | 0.384 | pvalb1 | -0.166 |
| XLOC-042222 | 0.383 | CU467822.1 | -0.164 |
| BX005254.3 | 0.379 | pvalb2 | -0.163 |
| noto | 0.376 | fabp3 | -0.157 |
| BX001014.2 | 0.375 | tmsb | -0.155 |
| znfl2a | 0.363 | tmsb4x | -0.153 |
| XLOC-001603 | 0.335 | nova2 | -0.152 |
| npm1a | 0.334 | gapdhs | -0.148 |
| cxcr4a | 0.324 | actc1b | -0.147 |
| lrrc17 | 0.322 | gpm6ab | -0.146 |
| XLOC-032526 | 0.315 | hbbe1.3 | -0.146 |
| sall4 | 0.312 | hbae3 | -0.141 |
| asb11 | 0.311 | zgc:165461 | -0.140 |
| pou5f3 | 0.305 | stmn1b | -0.140 |
| XLOC-039121 | 0.301 | COX3 | -0.139 |
| dkc1 | 0.297 | elavl3 | -0.139 |
| nkx1.2la | 0.294 | CU634008.1 | -0.139 |
| nop58 | 0.294 | rnasekb | -0.138 |
| greb1 | 0.284 | gpm6bb | -0.136 |
| XLOC-001964 | 0.283 | myt1b | -0.135 |
| nid2a | 0.281 | ppdpfb | -0.135 |
| mllt3 | 0.280 | si:dkeyp-75h12.5 | -0.134 |
| cx43.4 | 0.280 | vim | -0.132 |
| nhp2 | 0.277 | slc1a2b | -0.127 |
| snu13b | 0.276 | gnao1a | -0.124 |
| arid3b | 0.275 | atp1a1b | -0.123 |
| lamb1a | 0.273 | mylpfa | -0.122 |
| foxd5 | 0.271 | hbae1.1 | -0.120 |
| fbl | 0.270 | fez1 | -0.119 |