"NK1 transcription factor related 2-like,a"
ZFIN 
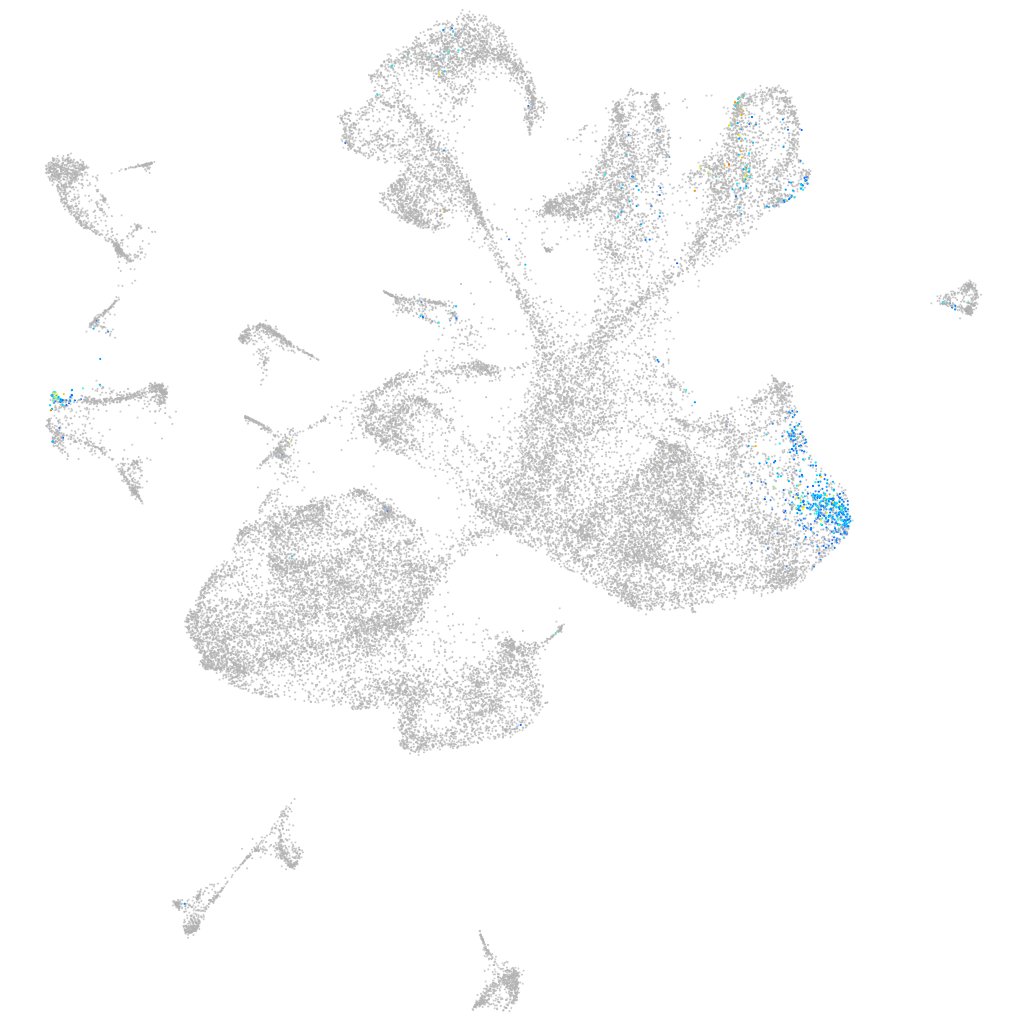
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm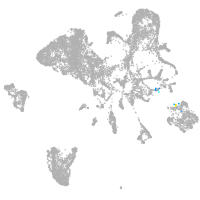 |
axial mesoderm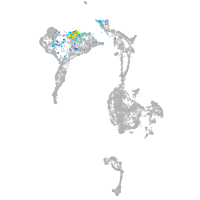 |
muscle 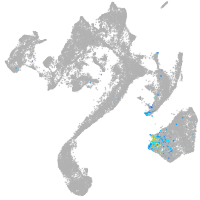 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural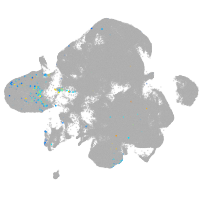 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| noto | 0.409 | fabp3 | -0.147 |
| sp5l | 0.406 | marcksl1b | -0.118 |
| chrd | 0.357 | tmsb4x | -0.115 |
| pou5f3 | 0.357 | fabp7a | -0.101 |
| fgf8a | 0.344 | CR383676.1 | -0.095 |
| cxcr4a | 0.300 | mdka | -0.094 |
| cdx4 | 0.299 | CU467822.1 | -0.092 |
| nradd | 0.294 | gpm6aa | -0.088 |
| pnx | 0.293 | zgc:165461 | -0.088 |
| foxd5 | 0.288 | gpm6bb | -0.087 |
| apela | 0.275 | atp1a1b | -0.083 |
| lrrc17 | 0.275 | vim | -0.081 |
| hspb1 | 0.250 | gfap | -0.079 |
| XLOC-042222 | 0.237 | CU634008.1 | -0.077 |
| cdx1a | 0.229 | si:ch73-21g5.7 | -0.075 |
| hoxd10a | 0.224 | pvalb1 | -0.074 |
| znfl2a | 0.215 | cd82a | -0.071 |
| BX005254.3 | 0.212 | atp1b4 | -0.070 |
| dnase1l4.1 | 0.204 | nova2 | -0.069 |
| hoxb10a | 0.202 | si:ch211-251b21.1 | -0.068 |
| hoxd12a | 0.191 | sparc | -0.067 |
| cpm | 0.189 | pou3f3b | -0.066 |
| lamb1a | 0.179 | celf2 | -0.066 |
| sall4 | 0.177 | pvalb2 | -0.066 |
| arid3b | 0.169 | slc1a2b | -0.065 |
| BX927258.1 | 0.169 | tmem47 | -0.064 |
| aebp1 | 0.169 | plp1a | -0.064 |
| kif26ab | 0.168 | nr2f1a | -0.064 |
| mllt3 | 0.167 | cspg5a | -0.063 |
| her3 | 0.167 | CR848047.1 | -0.063 |
| notum1a | 0.166 | marcksl1a | -0.063 |
| sp9 | 0.164 | fgfr3 | -0.063 |
| tbxta | 0.163 | actc1b | -0.063 |
| BX001014.2 | 0.160 | meis1b | -0.062 |
| znfl1k | 0.159 | COX3 | -0.060 |