nidogen 2a (osteonidogen)
ZFIN 
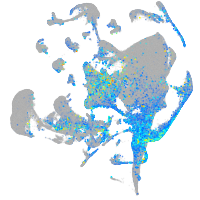
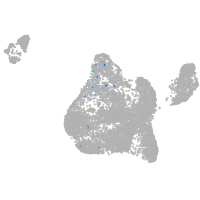
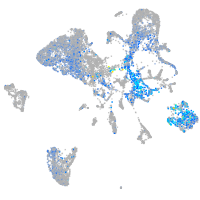
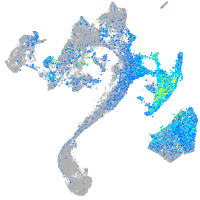
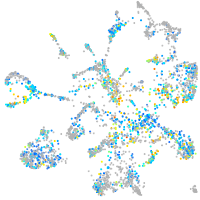
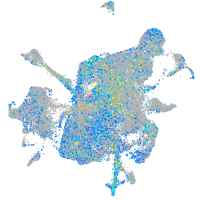
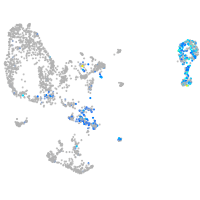
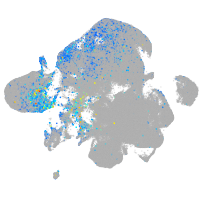
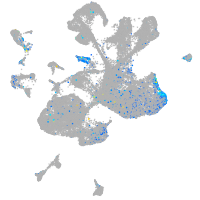
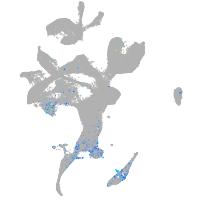
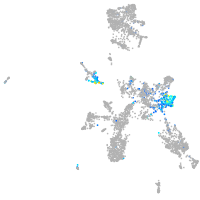
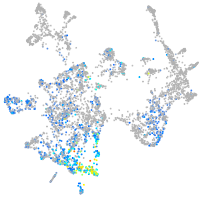
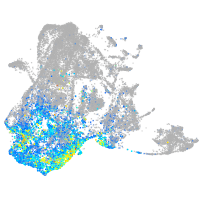
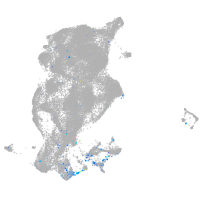
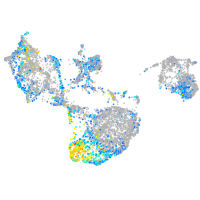
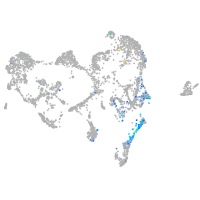
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoc1 | 0.519 | fabp3 | -0.406 |
| cx43.4 | 0.495 | actc1b | -0.361 |
| msgn1 | 0.494 | ckma | -0.339 |
| itm2cb | 0.492 | bhmt | -0.338 |
| fn1b | 0.480 | ckmb | -0.337 |
| her7 | 0.477 | atp2a1 | -0.334 |
| hes6 | 0.476 | ak1 | -0.322 |
| pcdh8 | 0.475 | pabpc4 | -0.319 |
| her1 | 0.467 | si:dkey-16p21.8 | -0.317 |
| apoeb | 0.438 | mylpfa | -0.313 |
| si:ch73-281n10.2 | 0.422 | eno1a | -0.312 |
| tbx16l | 0.420 | gamt | -0.310 |
| efnb2b | 0.418 | gapdh | -0.306 |
| tbx6 | 0.416 | pvalb1 | -0.305 |
| hoxb7a | 0.401 | tmem38a | -0.303 |
| myf5 | 0.401 | aldoab | -0.302 |
| her12 | 0.398 | pvalb2 | -0.300 |
| hoxb10a | 0.396 | eef1da | -0.299 |
| draxin | 0.393 | mylz3 | -0.294 |
| syncrip | 0.392 | nme2b.2 | -0.294 |
| hmga1a | 0.389 | acta1b | -0.292 |
| BX005254.3 | 0.389 | tnnc2 | -0.292 |
| dlc | 0.386 | ldb3b | -0.291 |
| CABZ01080702.1 | 0.386 | si:ch73-367p23.2 | -0.289 |
| khdrbs1a | 0.384 | myl1 | -0.289 |
| tspan7 | 0.383 | tpi1b | -0.288 |
| h2afvb | 0.380 | neb | -0.285 |
| gnaia | 0.379 | tnnt3a | -0.285 |
| aplnra | 0.372 | ank1a | -0.281 |
| cdh2 | 0.369 | actn3a | -0.276 |
| hnrnpaba | 0.366 | myom1a | -0.275 |
| notch1a | 0.366 | CABZ01078594.1 | -0.274 |
| h3f3d | 0.365 | sparc | -0.273 |
| tbx16 | 0.364 | srl | -0.272 |
| hoxc3a | 0.364 | tpma | -0.272 |