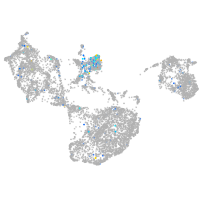
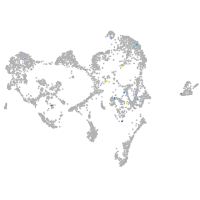
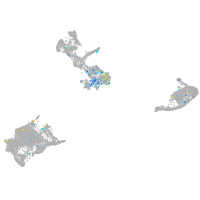
LIM homeobox 1a
ZFIN 
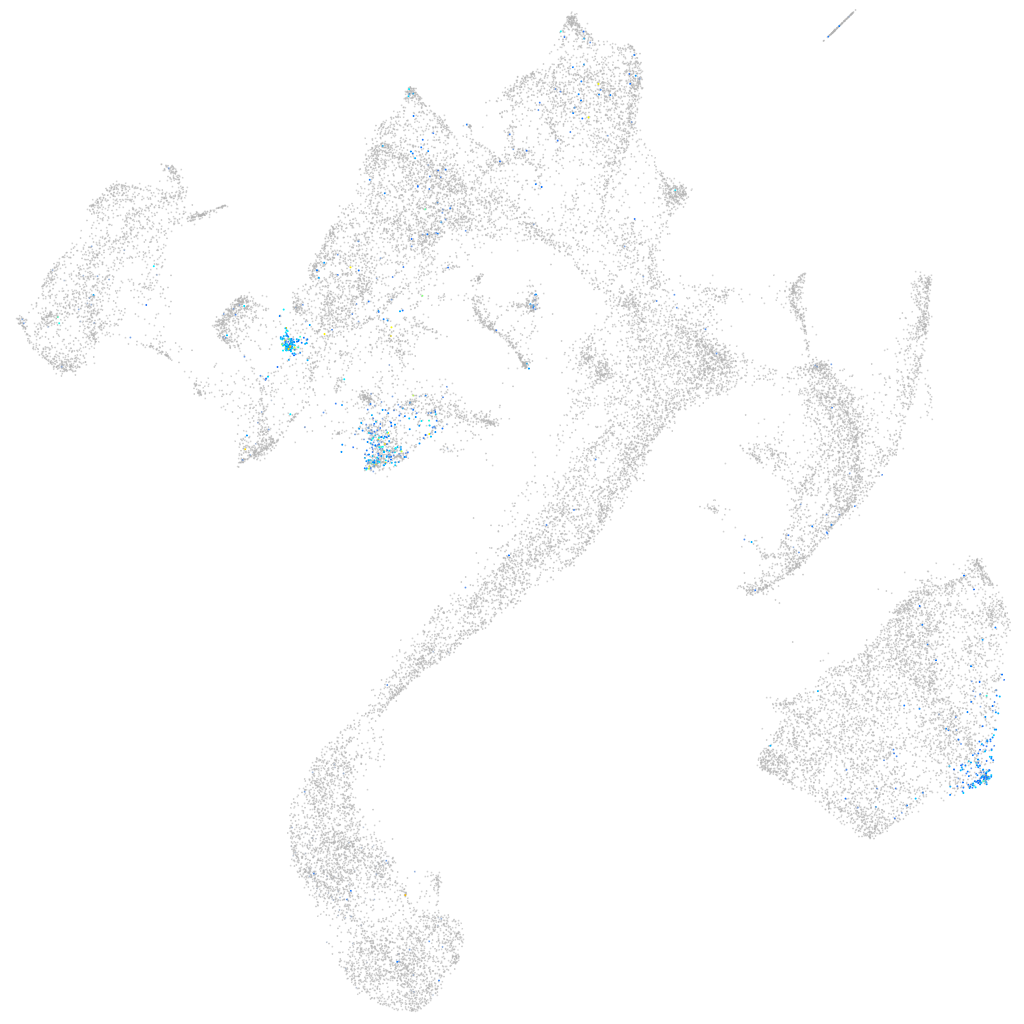
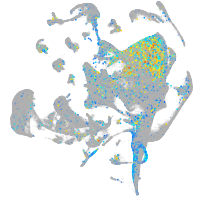
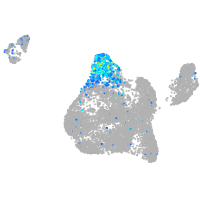
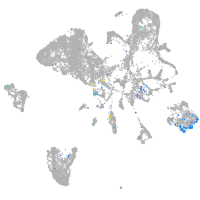
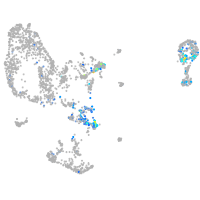
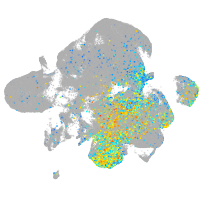
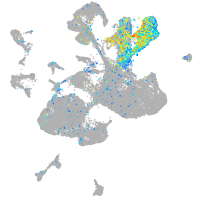
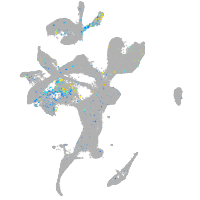
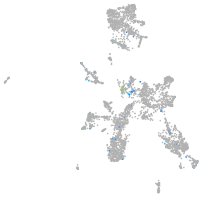
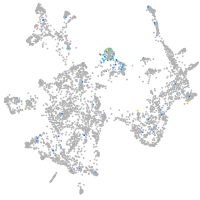
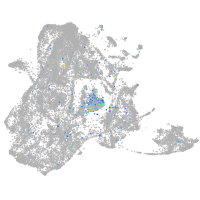
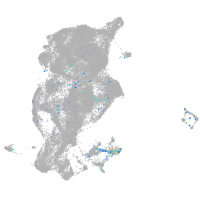
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ca4a | 0.236 | tuba8l2 | -0.098 |
| elavl4 | 0.215 | rplp0 | -0.064 |
| stmn1b | 0.212 | fn1b | -0.064 |
| elavl3 | 0.196 | cox6b2 | -0.061 |
| pitx2 | 0.196 | rpl6 | -0.060 |
| gata3 | 0.191 | BX001014.2 | -0.059 |
| gng3 | 0.189 | hoxb7a | -0.058 |
| nova2 | 0.189 | hoxc3a | -0.057 |
| stxbp1a | 0.188 | plp2 | -0.057 |
| gpm6aa | 0.188 | ttn.1 | -0.055 |
| sncb | 0.188 | zgc:92429 | -0.055 |
| zgc:65894 | 0.187 | meox1 | -0.055 |
| rtn1b | 0.186 | rps27a | -0.055 |
| ckbb | 0.184 | optc | -0.055 |
| stx1b | 0.184 | hoxc8a | -0.055 |
| fez1 | 0.179 | si:ch211-131k2.3 | -0.055 |
| stmn2a | 0.178 | tpma | -0.054 |
| emx1 | 0.177 | XLOC-042222 | -0.053 |
| tmsb2 | 0.175 | BX276101.1 | -0.053 |
| myt1b | 0.173 | srl | -0.052 |
| slc32a1 | 0.173 | efemp2a | -0.052 |
| tuba1c | 0.170 | XLOC-042229 | -0.052 |
| nsg2 | 0.170 | hsp90aa1.1 | -0.052 |
| lmx1ba | 0.170 | rpl8 | -0.052 |
| vamp2 | 0.167 | rpl10a | -0.052 |
| lhx5 | 0.167 | txlnbb | -0.051 |
| fscn1a | 0.166 | aldoab | -0.051 |
| cdk5r1b | 0.165 | COX5B | -0.051 |
| gabrb4 | 0.161 | uqcrh | -0.051 |
| fkbp1ab | 0.161 | rps26l | -0.051 |
| slc6a1b | 0.160 | tcf15 | -0.051 |
| atp6v0cb | 0.154 | hoxc6b | -0.050 |
| scrt1a | 0.154 | arid3c | -0.050 |
| snap25a | 0.154 | zgc:101853 | -0.050 |
| dpysl3 | 0.153 | actn3b | -0.050 |