FERM domain containing 7
ZFIN 
Other cell groups


all cells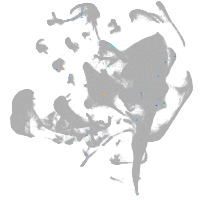 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 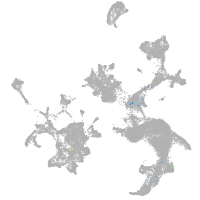 |
pronephros 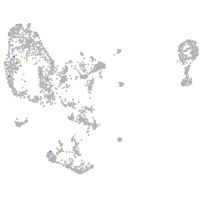 |
neural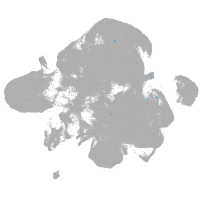 |
spinal cord / glia |
eye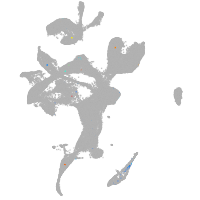 |
taste / olfactory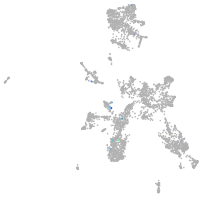 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous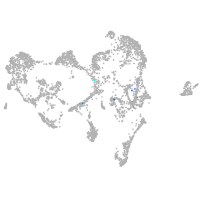 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cryba1l2 | 0.118 | aldob | -0.013 |
| gja8a | 0.118 | zgc:162730 | -0.013 |
| cx23 | 0.107 | fosab | -0.012 |
| BX548032.3 | 0.104 | ier2b | -0.012 |
| mipa | 0.103 | sparc | -0.012 |
| nags | 0.103 | ahnak | -0.011 |
| gja8b | 0.102 | btg2 | -0.011 |
| cps1 | 0.101 | cebpd | -0.011 |
| lim2.4 | 0.101 | ckap4 | -0.011 |
| bfsp2 | 0.100 | aldh9a1a.1 | -0.010 |
| lim2.3 | 0.098 | cfl1l | -0.010 |
| mipb | 0.094 | ctsla | -0.010 |
| crybb1l1 | 0.093 | cyt1 | -0.010 |
| lim2.1 | 0.093 | cyt1l | -0.010 |
| crybb1l3 | 0.092 | krt5 | -0.010 |
| gpx9 | 0.092 | mdka | -0.010 |
| BFSP1 | 0.091 | XLOC-044191 | -0.010 |
| cryba2a | 0.091 | mfap2 | -0.010 |
| crybb1l2 | 0.090 | anxa1a | -0.009 |
| cryba1b | 0.086 | apoeb | -0.009 |
| gja3 | 0.086 | arhgef5 | -0.009 |
| capn3a | 0.085 | boc | -0.009 |
| crygmx | 0.085 | cavin2b | -0.009 |
| crygmxl2 | 0.085 | cd63 | -0.009 |
| endou2 | 0.085 | cebpb | -0.009 |
| cryba1l1 | 0.084 | col1a1a | -0.009 |
| crybb1 | 0.082 | col1a2 | -0.009 |
| crygn2 | 0.080 | fstl1b | -0.009 |
| cryba2b | 0.078 | gsta.1 | -0.009 |
| cryba4 | 0.075 | jdp2b | -0.009 |
| prox2 | 0.073 | junba | -0.009 |
| grifin | 0.072 | krt17 | -0.009 |
| nrl | 0.072 | krt4 | -0.009 |
| otc | 0.071 | pfn1 | -0.009 |
| crygm2d13 | 0.069 | pmp22a | -0.009 |




