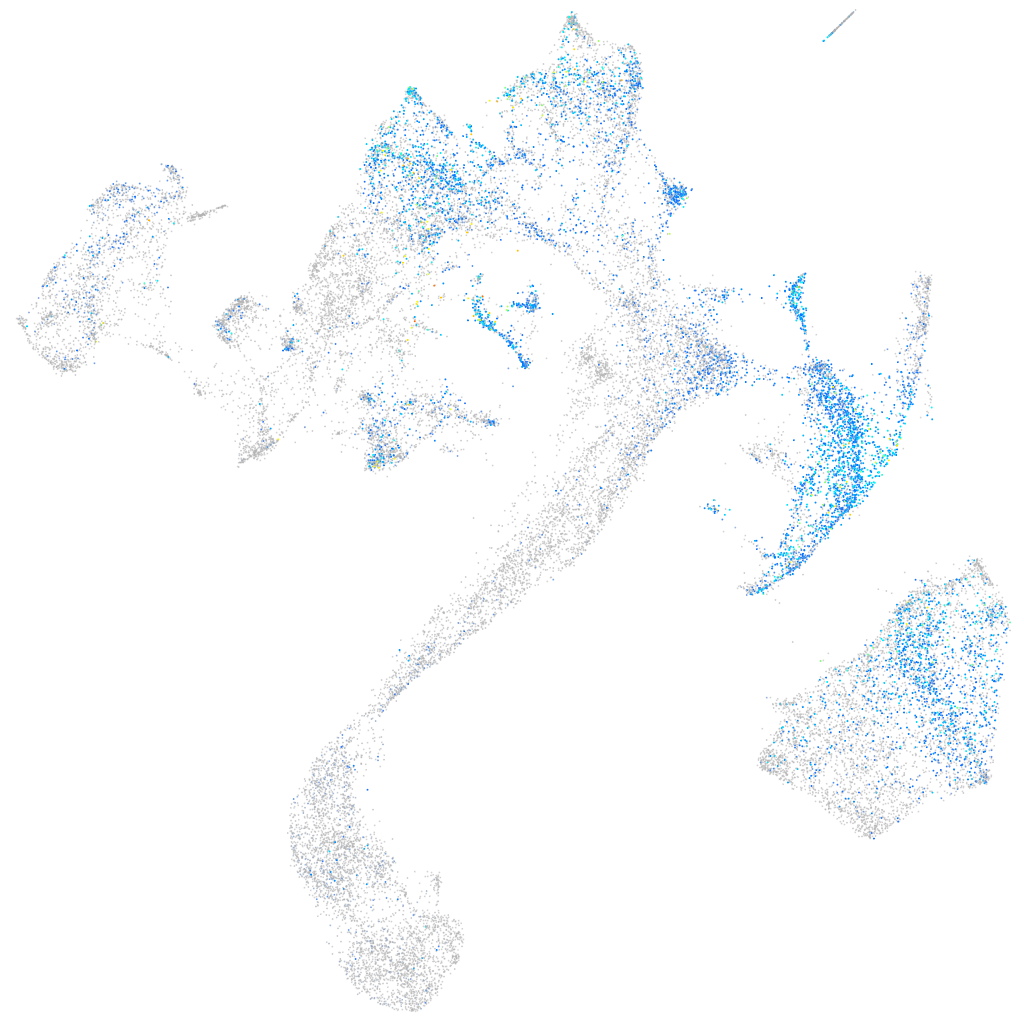
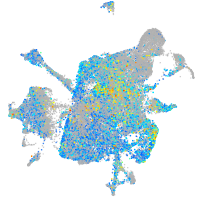
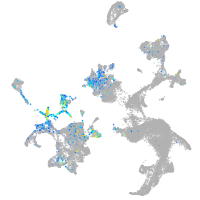
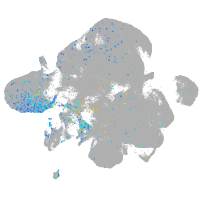
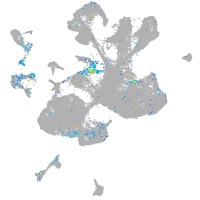
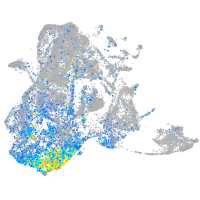
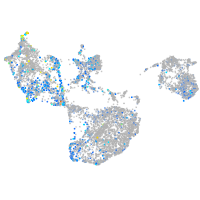
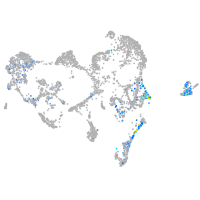
fibronectin 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msgn1 | 0.341 | actc1b | -0.262 |
| her1 | 0.337 | ttn.2 | -0.256 |
| itm2cb | 0.319 | pabpc4 | -0.252 |
| nid2a | 0.307 | tmem38a | -0.241 |
| her7 | 0.305 | aldoab | -0.237 |
| tbx16l | 0.303 | ak1 | -0.236 |
| ism1 | 0.298 | klhl41b | -0.233 |
| pcdh8 | 0.296 | atp2a1 | -0.232 |
| cx43.4 | 0.290 | ckmb | -0.230 |
| hoxb10a | 0.280 | ckma | -0.228 |
| tspan7 | 0.273 | ttn.1 | -0.225 |
| ephb3 | 0.272 | tnnc2 | -0.223 |
| hes6 | 0.267 | myl1 | -0.219 |
| cdh2 | 0.265 | acta1b | -0.218 |
| efnb2b | 0.262 | zgc:101853 | -0.218 |
| hoxd10a | 0.258 | CABZ01078594.1 | -0.217 |
| ubc | 0.256 | mybphb | -0.217 |
| apoc1 | 0.251 | desma | -0.216 |
| her12 | 0.250 | gapdh | -0.215 |
| lpar1 | 0.250 | ldb3a | -0.214 |
| CABZ01080702.1 | 0.247 | srl | -0.214 |
| gnaia | 0.246 | tpma | -0.213 |
| hsp90ab1 | 0.243 | neb | -0.213 |
| tbx6 | 0.242 | txlnbb | -0.213 |
| foxc1a | 0.242 | cox17 | -0.212 |
| fkbp7 | 0.238 | gatm | -0.212 |
| LO016987.2 | 0.236 | eno1a | -0.211 |
| fzd10 | 0.233 | mylpfa | -0.211 |
| asph | 0.233 | cav3 | -0.209 |
| dlc | 0.232 | gamt | -0.208 |
| BX005254.3 | 0.232 | ldb3b | -0.204 |
| ptmab | 0.232 | acta1a | -0.201 |
| metrnla | 0.230 | atp1a2a | -0.197 |
| greb1 | 0.230 | atp5if1b | -0.197 |
| raraa | 0.230 | si:ch73-367p23.2 | -0.197 |