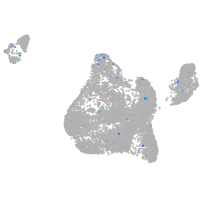
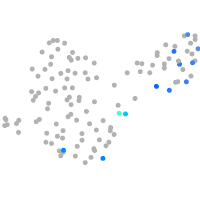
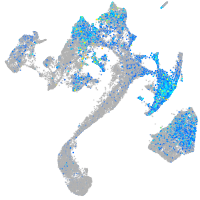
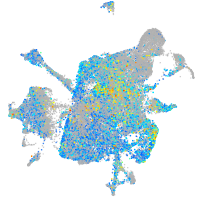
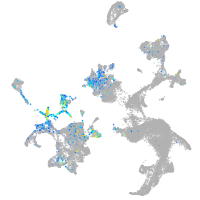
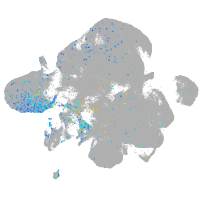
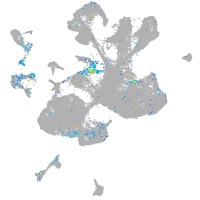
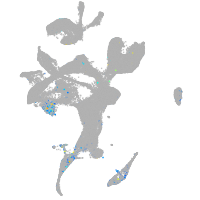
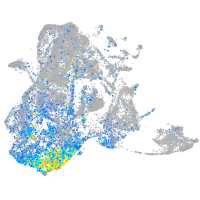
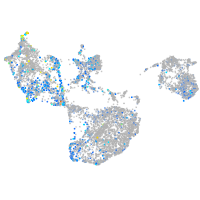
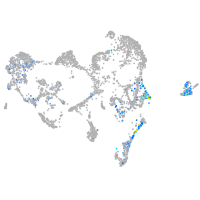
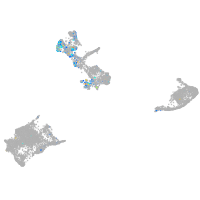
fibronectin 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.310 | atp5f1b | -0.267 |
| cxcl12a | 0.308 | eno3 | -0.257 |
| apoc1 | 0.304 | atp1b1a | -0.249 |
| hspb1 | 0.299 | si:ch211-139a5.9 | -0.245 |
| cdx4 | 0.292 | suclg1 | -0.243 |
| gck | 0.290 | atp5mc1 | -0.243 |
| apoeb | 0.287 | atp5mc3b | -0.241 |
| nid2a | 0.279 | rpl37 | -0.236 |
| scn4ab | 0.278 | rps10 | -0.229 |
| gfra4b | 0.277 | atp5f1e | -0.226 |
| BX927327.1 | 0.273 | atp5l | -0.224 |
| calca | 0.272 | aldh6a1 | -0.221 |
| id3 | 0.266 | cox5ab | -0.221 |
| qkia | 0.263 | zgc:114188 | -0.219 |
| fkbp7 | 0.262 | atp5pb | -0.219 |
| nr0b1 | 0.262 | COX3 | -0.218 |
| wu:fb97g03 | 0.260 | atp5f1d | -0.216 |
| ilf3b | 0.258 | vdac3 | -0.215 |
| akap12b | 0.256 | si:dkey-16p21.8 | -0.214 |
| ved | 0.256 | ldhba | -0.214 |
| cdx1a | 0.254 | cox8a | -0.214 |
| bambia | 0.248 | rps17 | -0.213 |
| anp32b | 0.247 | cdh17 | -0.213 |
| nucks1a | 0.246 | atp5fa1 | -0.212 |
| msna | 0.246 | idh2 | -0.211 |
| dnmt3bb.2 | 0.244 | ndufb10 | -0.210 |
| nop58 | 0.242 | atp1a1a.4 | -0.209 |
| cx39.4 | 0.241 | cox6c | -0.208 |
| hnrnpabb | 0.240 | atp5mf | -0.208 |
| ncl | 0.238 | atp5po | -0.207 |
| entpd1 | 0.236 | atp5if1b | -0.207 |
| blf | 0.235 | mdh1aa | -0.207 |
| srrm1 | 0.235 | mt-nd4 | -0.206 |
| syncrip | 0.234 | cox6b1 | -0.205 |
| ppig | 0.233 | cox6a1 | -0.205 |