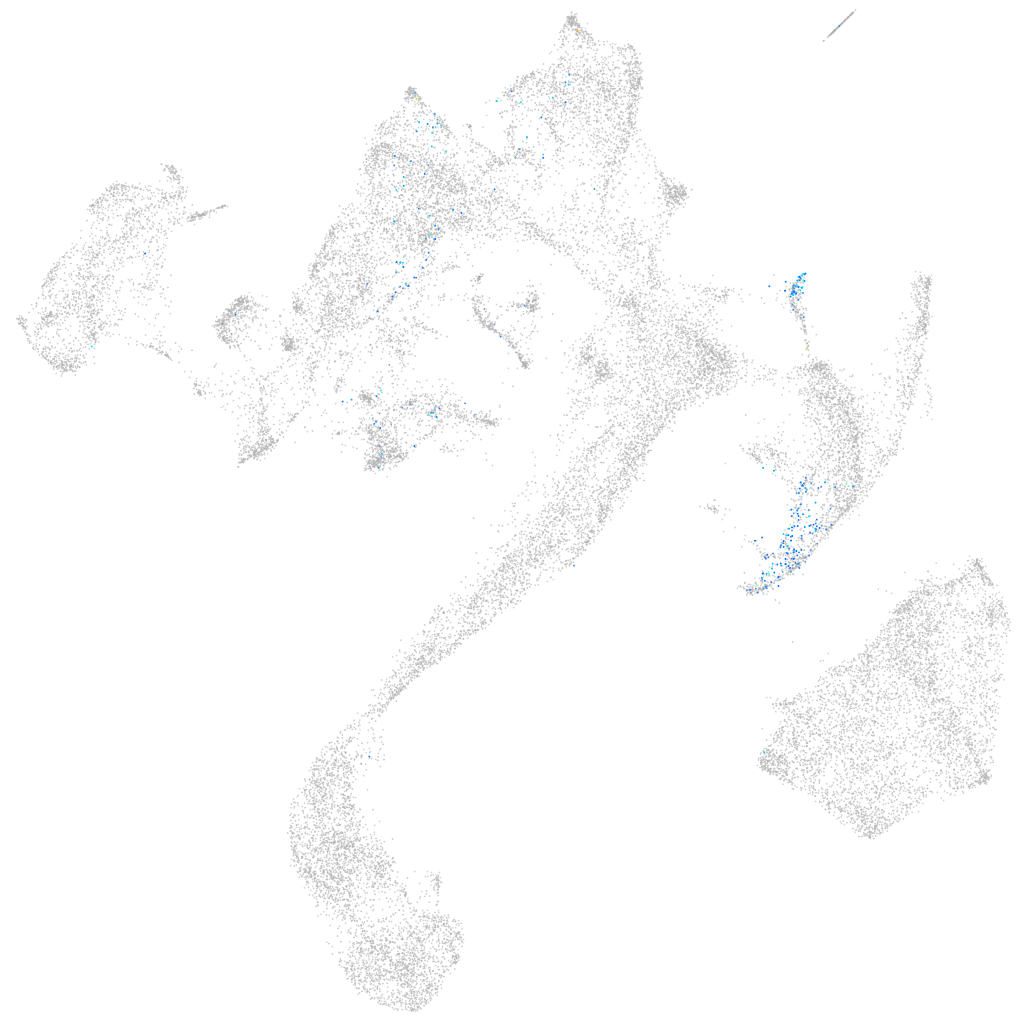
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-004335 | 0.301 | ttn.1 | -0.066 |
| LOC101887111 | 0.258 | hsp90aa1.1 | -0.064 |
| hoxa13b | 0.238 | ttn.2 | -0.060 |
| hoxc13b | 0.201 | fxr2 | -0.058 |
| fgf3 | 0.197 | zgc:92429 | -0.056 |
| gdf11 | 0.188 | actc1b | -0.056 |
| hoxd12a | 0.188 | ak1 | -0.054 |
| hoxc13a | 0.178 | aldoab | -0.053 |
| tagln3b | 0.152 | tmem38a | -0.053 |
| hoxa13a | 0.151 | gapdh | -0.053 |
| hoxd10a | 0.146 | mybphb | -0.052 |
| itm2cb | 0.138 | pabpc4 | -0.052 |
| thbs2a | 0.138 | klhl41b | -0.051 |
| hoxd13a | 0.138 | atp2a1 | -0.051 |
| hoxb10a | 0.137 | ckmb | -0.051 |
| cyp26a1 | 0.134 | actc1a | -0.050 |
| zgc:162939 | 0.134 | tnnc2 | -0.050 |
| hoxc12b | 0.134 | CABZ01078594.1 | -0.050 |
| hoxc11a | 0.130 | acta1a | -0.050 |
| bmp2a | 0.129 | zgc:92518 | -0.050 |
| her12 | 0.129 | desma | -0.050 |
| tll1 | 0.128 | zgc:101853 | -0.049 |
| her15.1 | 0.127 | ldb3a | -0.049 |
| zgc:162707 | 0.127 | ckma | -0.049 |
| esrrga | 0.125 | gatm | -0.048 |
| hoxa11a | 0.124 | srl | -0.048 |
| fgf10a | 0.123 | rbfox1l | -0.048 |
| vimr2 | 0.123 | tpma | -0.048 |
| hoxb13a | 0.121 | acta1b | -0.047 |
| sall4 | 0.120 | neb | -0.047 |
| fgf8a | 0.119 | chchd10 | -0.047 |
| her1 | 0.118 | actn3b | -0.047 |
| XLOC-003690 | 0.117 | cav3 | -0.046 |
| si:cabz01036006.1 | 0.117 | txlnbb | -0.046 |
| her7 | 0.117 | smyd1b | -0.046 |