zgc:165582
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis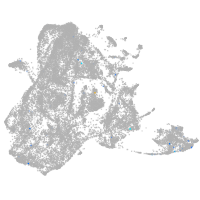 |
periderm |
fin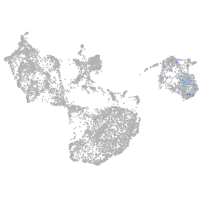 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grinaa | 0.796 | hmga1a | -0.304 |
| XLOC-016430 | 0.777 | dnajc1 | -0.291 |
| nlgn4xa | 0.751 | hnrnpub | -0.267 |
| LOC101883509 | 0.750 | pabpc1a | -0.247 |
| mir22a | 0.750 | ctsla | -0.231 |
| plpp2a | 0.750 | hnrnpabb | -0.228 |
| wasf3a | 0.750 | NC-002333.4 | -0.221 |
| XLOC-016167 | 0.750 | h1m | -0.220 |
| trim35-33 | 0.709 | hnrnph1l | -0.220 |
| nr2f5 | 0.698 | srrt | -0.215 |
| fcer1gl | 0.693 | rps6 | -0.211 |
| tns1b | 0.693 | top1l | -0.211 |
| adra2da | 0.677 | hnrnpa1a | -0.209 |
| AL928808.1 | 0.677 | acin1a | -0.208 |
| atp1a1a.4 | 0.677 | chaf1a | -0.208 |
| BX000451.1 | 0.677 | hnrnpa1b | -0.208 |
| BX649398.1 | 0.677 | stm | -0.205 |
| BX649434.2 | 0.677 | hnrnpl | -0.205 |
| CR855277.3 | 0.677 | ccna2 | -0.202 |
| CT009487.2 | 0.677 | cdk11b | -0.196 |
| FO704607.1 | 0.677 | acin1b | -0.196 |
| gpr61 | 0.677 | nkap | -0.195 |
| hoxa1a | 0.677 | sf3b4 | -0.195 |
| hoxa5a | 0.677 | nucks1a | -0.193 |
| hoxb1a | 0.677 | hmgb2a | -0.191 |
| hoxb8b | 0.677 | zgc:110425 | -0.190 |
| LOC101886896 | 0.677 | sf3b2 | -0.188 |
| LOC103908766 | 0.677 | sart1 | -0.186 |
| LOC108192140 | 0.677 | dnaja2a | -0.186 |
| LOC110438192 | 0.677 | odc1 | -0.183 |
| LOC110438604 | 0.677 | cd2bp2 | -0.182 |
| loxl5b | 0.677 | stmn1a | -0.182 |
| map3k19 | 0.677 | slc16a3 | -0.182 |
| mir301b | 0.677 | syncrip | -0.181 |
| ms4a17a.17 | 0.677 | hspb1 | -0.178 |




