Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 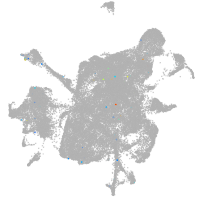 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye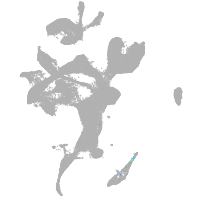 |
taste / olfactory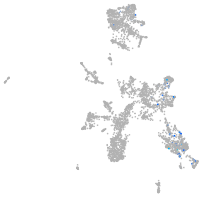 |
otic / lateral line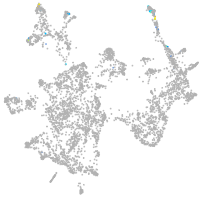 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU062645.1 | 0.070 | hmgb1b | -0.022 |
| zgc:195077 | 0.068 | si:ch211-288g17.3 | -0.022 |
| myl4 | 0.065 | gpm6aa | -0.021 |
| BX548032.3 | 0.063 | nova2 | -0.021 |
| cryba1l2 | 0.063 | marcksb | -0.020 |
| emb | 0.062 | si:ch211-137a8.4 | -0.020 |
| capn3a | 0.061 | h2afva | -0.018 |
| mustn1b | 0.060 | seta | -0.017 |
| myoz2b | 0.058 | hnrnpa0a | -0.016 |
| smpx | 0.058 | nono | -0.016 |
| lim2.5 | 0.058 | nucks1a | -0.016 |
| xirp1 | 0.057 | smarce1 | -0.016 |
| ryr1a | 0.056 | alyref | -0.015 |
| prx | 0.055 | cbx3a | -0.015 |
| smyhc2 | 0.054 | chd4a | -0.015 |
| si:dkey-164f24.2 | 0.053 | cspg5a | -0.015 |
| CU179656.2 | 0.052 | hdac1 | -0.015 |
| mylk4a | 0.052 | hnrnpa1a | -0.015 |
| klhl41a | 0.051 | mdkb | -0.015 |
| trarg1b | 0.051 | rbbp4 | -0.015 |
| hspb11 | 0.050 | snrpf | -0.015 |
| krt97 | 0.050 | sox11b | -0.015 |
| gpx9 | 0.049 | sumo3b | -0.015 |
| lmod3 | 0.049 | top1l | -0.015 |
| mipa | 0.049 | zc4h2 | -0.015 |
| twf2a | 0.049 | apex1 | -0.014 |
| zgc:158296 | 0.049 | hmgb3a | -0.014 |
| CR356246.1 | 0.048 | hnrnpa1b | -0.014 |
| hhatlb | 0.048 | hnrnph1l | -0.014 |
| lim2.4 | 0.048 | hnrnpub | -0.014 |
| moxd1l | 0.048 | ilf3b | -0.014 |
| mybpc1 | 0.048 | rcc2 | -0.014 |
| nrap | 0.048 | setb | -0.014 |
| p2rx3a | 0.048 | atrx | -0.013 |
| rnf207b | 0.048 | baz1b | -0.013 |




