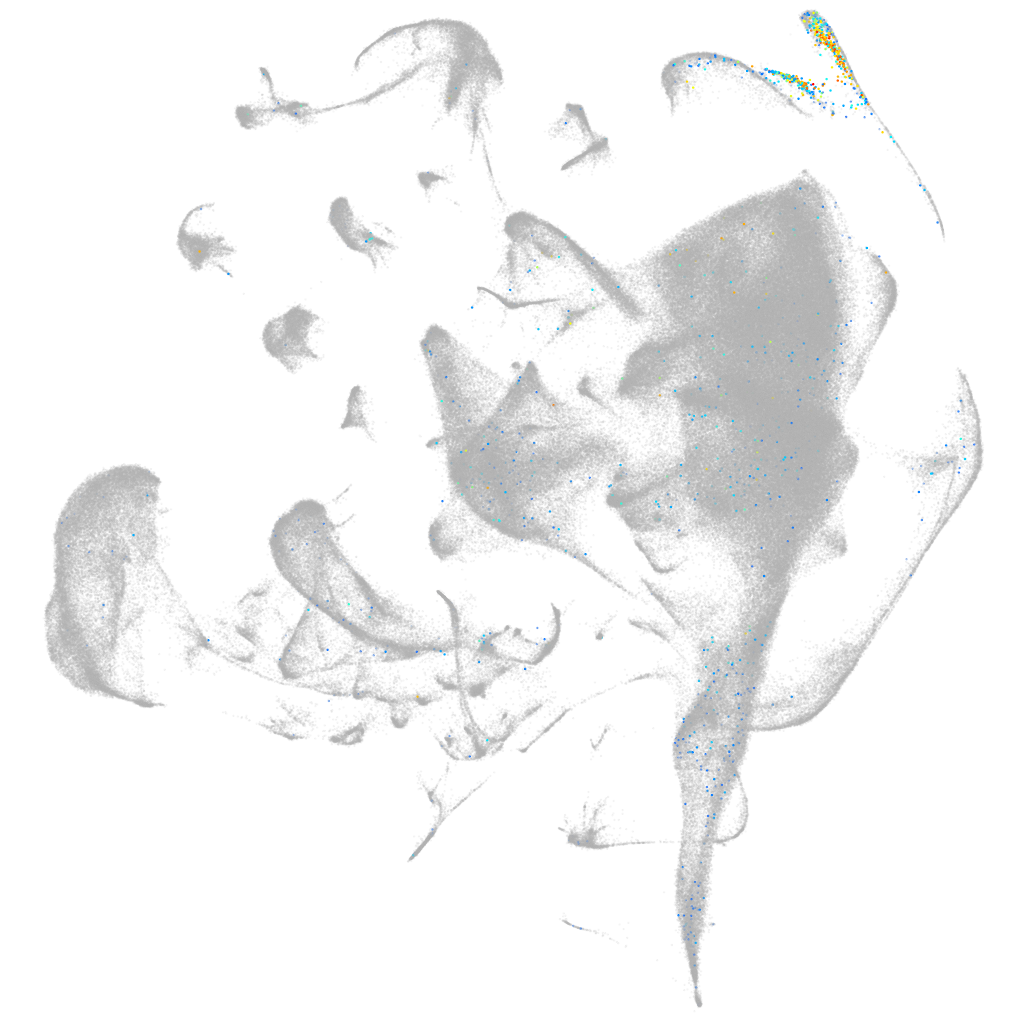myosin binding protein C1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mustn1b | 0.237 | gpm6ab | -0.030 |
| ryr1a | 0.225 | gapdhs | -0.028 |
| myl4 | 0.223 | atp6v0cb | -0.026 |
| lmod2b | 0.215 | rnasekb | -0.026 |
| MYL2 | 0.213 | ckbb | -0.024 |
| hspb11 | 0.203 | gpm6aa | -0.024 |
| obscna | 0.201 | rtn1b | -0.024 |
| flnca | 0.200 | sncb | -0.024 |
| fhl5 | 0.196 | atp6ap2 | -0.023 |
| tnni1d | 0.194 | cadm3 | -0.023 |
| klhl41a | 0.193 | calm1a | -0.023 |
| zgc:172270 | 0.193 | atpv0e2 | -0.022 |
| zgc:86709 | 0.186 | sypb | -0.022 |
| rnf207b | 0.185 | atp1a3a | -0.021 |
| tnni2a.1 | 0.185 | calm3b | -0.021 |
| tnni2a.3 | 0.182 | cspg5a | -0.021 |
| AGBL1 | 0.180 | stx1b | -0.021 |
| LOC110440135 | 0.178 | vamp2 | -0.021 |
| si:rp71-17i16.4 | 0.177 | ywhaqb | -0.021 |
| mylk4a | 0.174 | atp6v1e1b | -0.020 |
| tpm2 | 0.173 | calr | -0.020 |
| myoz3a | 0.171 | elavl4 | -0.020 |
| casq2 | 0.170 | gng3 | -0.020 |
| pdlim3b | 0.170 | cpe | -0.019 |
| myoz2b | 0.169 | epb41a | -0.019 |
| smpx | 0.169 | nptna | -0.019 |
| CU681836.1 | 0.166 | olfm1b | -0.019 |
| smyhc2 | 0.166 | stmn1b | -0.019 |
| actn2b | 0.165 | sv2a | -0.019 |
| cav3 | 0.163 | tuba1c | -0.019 |
| tcap | 0.163 | syt1a | -0.019 |
| myh7 | 0.162 | aplp1 | -0.018 |
| pvalb7 | 0.162 | calm2a | -0.018 |
| cavin4a | 0.161 | nrxn1a | -0.018 |
| myhb | 0.161 | nsg2 | -0.018 |