"musculoskeletal, embryonic nuclear protein 1b"
ZFIN 
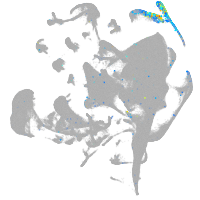
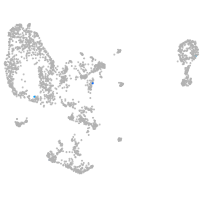
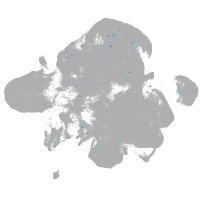
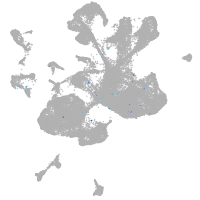
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ryr1a | 0.572 | ywhaqb | -0.071 |
| casq2 | 0.550 | gpm6aa | -0.069 |
| myoz2b | 0.545 | ckbb | -0.067 |
| cavin4a | 0.525 | gpm6ab | -0.067 |
| srl | 0.520 | rnasekb | -0.067 |
| zgc:158296 | 0.517 | marcksl1a | -0.067 |
| zgc:86709 | 0.513 | gapdhs | -0.066 |
| LO018250.1 | 0.506 | atp6v0cb | -0.065 |
| cavin4b | 0.502 | hmgb1b | -0.062 |
| cav3 | 0.496 | si:ch211-288g17.3 | -0.061 |
| hhatlb | 0.493 | tuba1c | -0.061 |
| desma | 0.489 | mdkb | -0.060 |
| CU633479.2 | 0.484 | nova2 | -0.060 |
| tnnc1b | 0.481 | ddx5 | -0.058 |
| tnni1c | 0.480 | gnb1b | -0.058 |
| hspb11 | 0.478 | chd4a | -0.057 |
| pgam2 | 0.477 | calm1a | -0.057 |
| klhl41a | 0.472 | atp6ap2 | -0.056 |
| nrap | 0.463 | calm3b | -0.056 |
| stac3 | 0.462 | atpv0e2 | -0.055 |
| apobec2a | 0.461 | atrx | -0.055 |
| trdn | 0.455 | h2afva | -0.055 |
| acta1a | 0.452 | pfn2 | -0.055 |
| CU681836.1 | 0.452 | tubb5 | -0.055 |
| myom1b | 0.452 | atp6v0ca | -0.054 |
| si:ch211-28p3.4 | 0.451 | cspg5a | -0.054 |
| smyhc2 | 0.451 | calr | -0.053 |
| actc1a | 0.450 | rtn1b | -0.053 |
| klhl43 | 0.447 | tmem258 | -0.053 |
| ldb3b | 0.444 | zc4h2 | -0.053 |
| myot | 0.444 | stmn1b | -0.052 |
| smpx | 0.444 | atp1a3a | -0.051 |
| sgca | 0.442 | sncb | -0.051 |
| aldoab | 0.440 | sypb | -0.051 |
| tmem182a | 0.439 | tmeff1b | -0.051 |