ATPase H+ transporting V0 subunit ca
ZFIN 
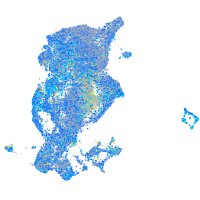
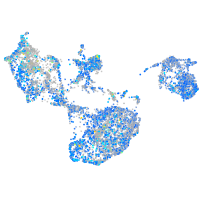
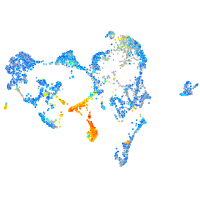
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1b | 0.173 | actc1b | -0.105 |
| sv2a | 0.170 | pvalb1 | -0.102 |
| atp6v0b | 0.165 | pvalb2 | -0.101 |
| atp6ap2 | 0.164 | tmod4 | -0.101 |
| atp6v1e1b | 0.163 | mylz3 | -0.091 |
| stx1b | 0.163 | hbbe2 | -0.090 |
| vat1 | 0.162 | ckma | -0.088 |
| gnb1b | 0.160 | ckmb | -0.085 |
| stmn2a | 0.160 | mylpfa | -0.085 |
| ywhag2 | 0.160 | neb | -0.082 |
| gdi1 | 0.158 | tnnt3b | -0.082 |
| tspan36 | 0.157 | actn3a | -0.081 |
| stxbp1a | 0.157 | apoa2 | -0.080 |
| sncb | 0.155 | ldb3b | -0.080 |
| dct | 0.155 | tnni2a.4 | -0.080 |
| atp6v1g1 | 0.154 | ttn.2 | -0.080 |
| tyrp1a | 0.153 | pgam2 | -0.080 |
| gnao1a | 0.151 | actn3b | -0.079 |
| slc45a2 | 0.151 | hbbe1.1 | -0.079 |
| pmela | 0.148 | si:ch211-250g4.3 | -0.079 |
| tspan10 | 0.148 | si:ch211-266g18.10 | -0.079 |
| vamp2 | 0.148 | si:dkey-16p21.8 | -0.079 |
| zgc:65894 | 0.148 | si:ch73-367p23.2 | -0.078 |
| atp6v0cb | 0.147 | tnnc2 | -0.078 |
| elavl4 | 0.147 | ttn.1 | -0.078 |
| sypa | 0.147 | aldoab | -0.077 |
| ywhaqb | 0.147 | casq1b | -0.077 |
| tyrp1b | 0.147 | myom1a | -0.077 |
| atp6v1h | 0.146 | pkmb | -0.077 |
| tyr | 0.146 | rbp4 | -0.077 |
| calr | 0.145 | atp2a1 | -0.077 |
| gap43 | 0.145 | cahz | -0.076 |
| dnajc5aa | 0.144 | casq2 | -0.076 |
| oca2 | 0.144 | hbae1.3 | -0.076 |
| syngr3a | 0.143 | hemgn | -0.076 |