ATPase H+ transporting V0 subunit ca
ZFIN 
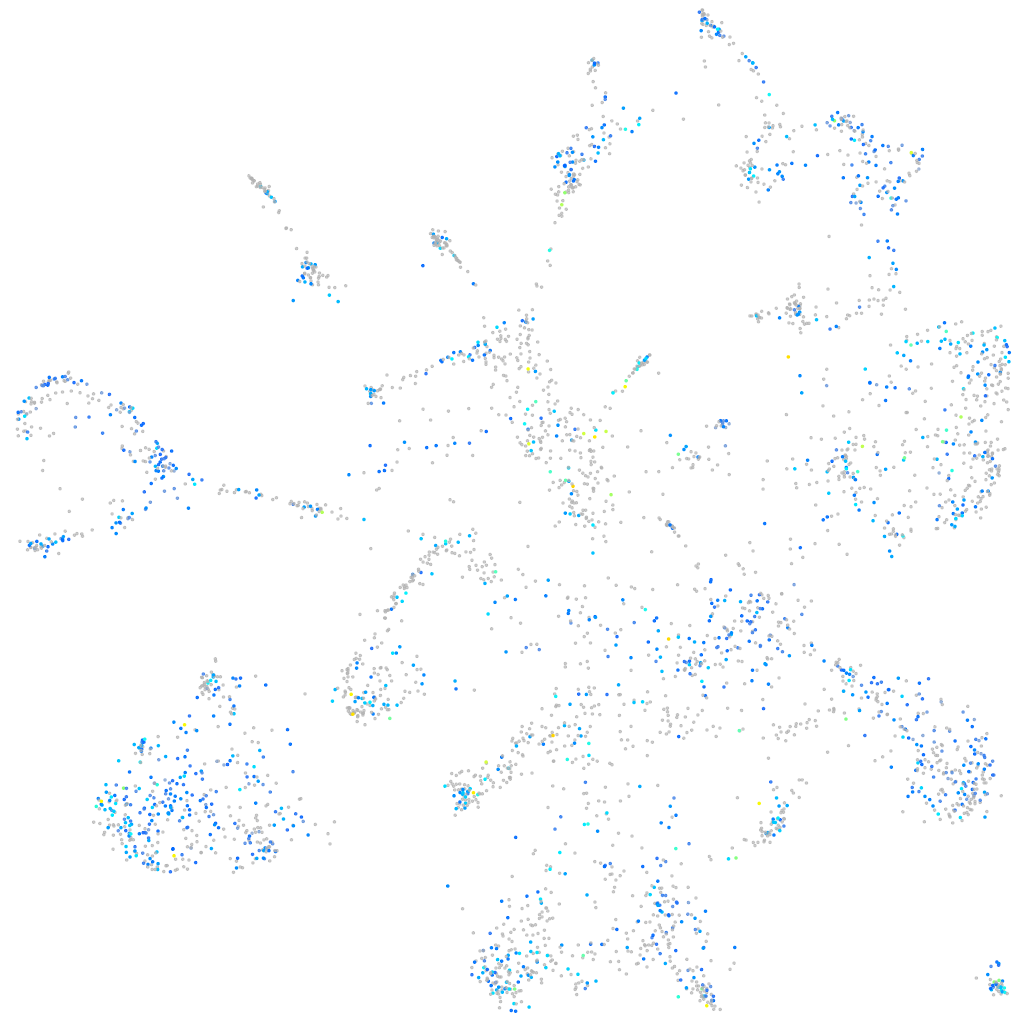
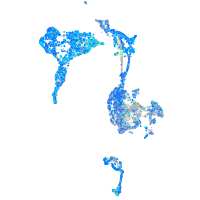
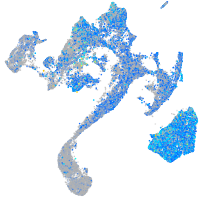
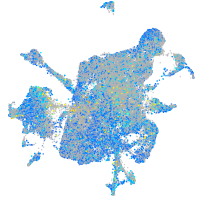
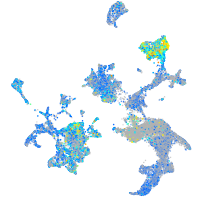
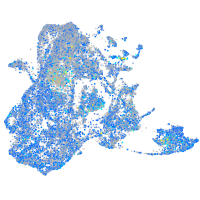
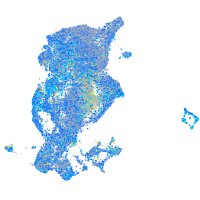
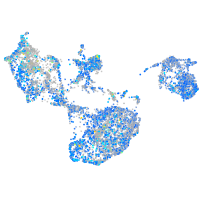
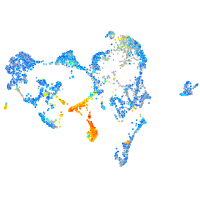
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ywhag2 | 0.146 | rplp1 | -0.138 |
| mt-nd4 | 0.145 | rps29 | -0.103 |
| sncb | 0.131 | rpl39 | -0.098 |
| mt-nd2 | 0.129 | rpl37 | -0.091 |
| ywhaqb | 0.129 | rbp4 | -0.083 |
| calr3b | 0.128 | pvalb2 | -0.082 |
| pfn2 | 0.127 | rps2 | -0.076 |
| hspa5 | 0.127 | pvalb1 | -0.075 |
| mt-cyb | 0.126 | prss59.2 | -0.073 |
| dad1 | 0.122 | rpl28 | -0.072 |
| slc32a1 | 0.122 | cebpd | -0.072 |
| fkbp9 | 0.121 | rpl36a | -0.071 |
| p4hb | 0.121 | ctrb1 | -0.070 |
| gabrb4 | 0.121 | rpl29 | -0.070 |
| pdia3 | 0.121 | si:dkey-151g10.6 | -0.069 |
| stxbp1a | 0.121 | rps21 | -0.069 |
| tuba1c | 0.121 | rpl19 | -0.066 |
| atp5mc3a | 0.121 | dap1b | -0.066 |
| ywhabl | 0.120 | rps27a | -0.066 |
| mt-atp6 | 0.120 | apoa1b | -0.065 |
| cdk5r1b | 0.120 | CELA1 (1 of many) | -0.064 |
| mt-nd1 | 0.119 | actc1b | -0.063 |
| elavl4 | 0.119 | ifitm1 | -0.063 |
| scamp5a | 0.118 | apoa2 | -0.062 |
| sv2a | 0.118 | rpl22 | -0.061 |
| gad2 | 0.117 | rplp0 | -0.060 |
| hnrnpa0a | 0.116 | myhz2 | -0.060 |
| ammecr1 | 0.116 | rps24 | -0.058 |
| spcs2 | 0.116 | rps19 | -0.057 |
| nova1 | 0.115 | rps20 | -0.057 |
| TRIM67 | 0.115 | rpl26 | -0.056 |
| gng3 | 0.114 | rpl38 | -0.056 |
| cirbpa | 0.113 | rps13 | -0.055 |
| atp1a3a | 0.113 | rpl24 | -0.054 |
| ap2m1a | 0.113 | rps14 | -0.054 |