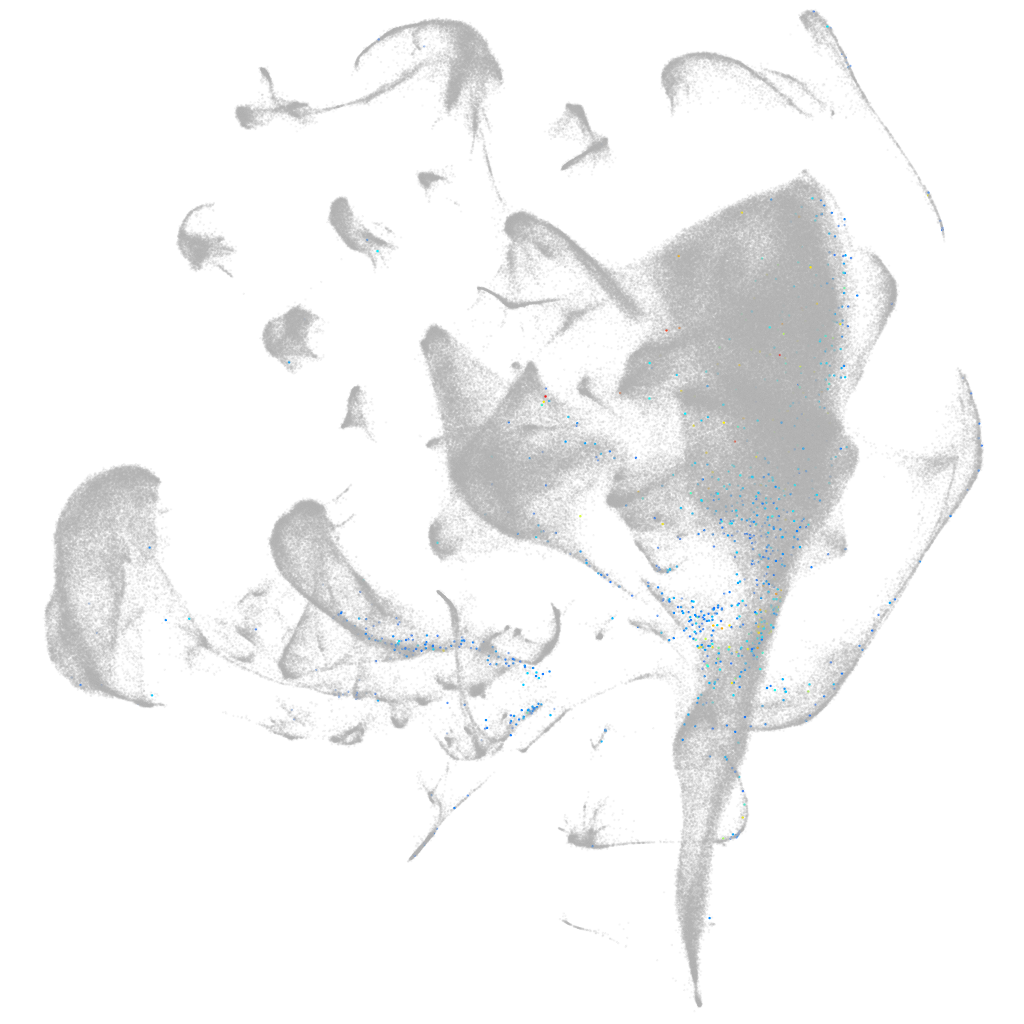
homeobox B1a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 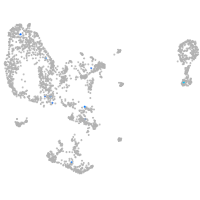 |
neural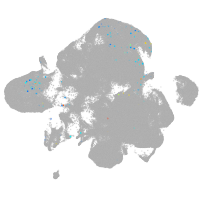 |
spinal cord / glia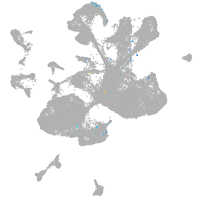 |
eye |
taste / olfactory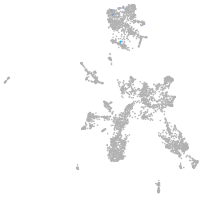 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous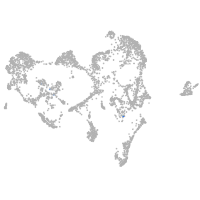 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxb1b | 0.072 | pvalb1 | -0.024 |
| ripply3 | 0.070 | pvalb2 | -0.023 |
| hoxb3a | 0.069 | eef1da | -0.022 |
| CABZ01075068.1 | 0.064 | ckbb | -0.018 |
| BX469925.3 | 0.062 | actc1b | -0.017 |
| meis3 | 0.055 | gapdhs | -0.017 |
| nr6a1a | 0.048 | icn | -0.017 |
| lin28a | 0.046 | krtt1c19e | -0.017 |
| hoxb2a | 0.045 | ahcy | -0.016 |
| greb1l | 0.043 | hbae1.1 | -0.016 |
| BX927258.1 | 0.042 | hbbe1.3 | -0.016 |
| CU571061.1 | 0.039 | icn2 | -0.016 |
| hspb1 | 0.038 | s100v2 | -0.016 |
| alcamb | 0.037 | anxa1a | -0.015 |
| dhrs3a | 0.035 | hbae3 | -0.015 |
| nr2f6b | 0.035 | krt4 | -0.015 |
| trim71 | 0.035 | krt5 | -0.015 |
| hoxc1a | 0.034 | rbp4 | -0.015 |
| igf2bp1 | 0.034 | col1a1a | -0.014 |
| nkx2.7 | 0.033 | cyt1 | -0.014 |
| irx7 | 0.032 | fbp1a | -0.014 |
| lsp1 | 0.032 | gpm6ab | -0.014 |
| meis1a | 0.032 | ndrg2 | -0.014 |
| npm1a | 0.032 | pycard | -0.014 |
| zfp36l1a | 0.032 | s100a10b | -0.014 |
| dhrs3b | 0.031 | si:ch211-105c13.3 | -0.014 |
| fstl1a | 0.031 | si:ch211-195b11.3 | -0.014 |
| meis1b | 0.031 | sod1 | -0.014 |
| ptges3b | 0.031 | tcnbb | -0.014 |
| apela | 0.030 | aep1 | -0.013 |
| hoxb5a | 0.030 | anxa2a | -0.013 |
| si:dkey-195m11.8 | 0.030 | atp5if1b | -0.013 |
| fbn2a.1 | 0.030 | gstp1 | -0.013 |
| meis2b | 0.030 | hbbe1.1 | -0.013 |
| nop58 | 0.029 | krt17 | -0.013 |