solute carrier family 4 member 2b
ZFIN 
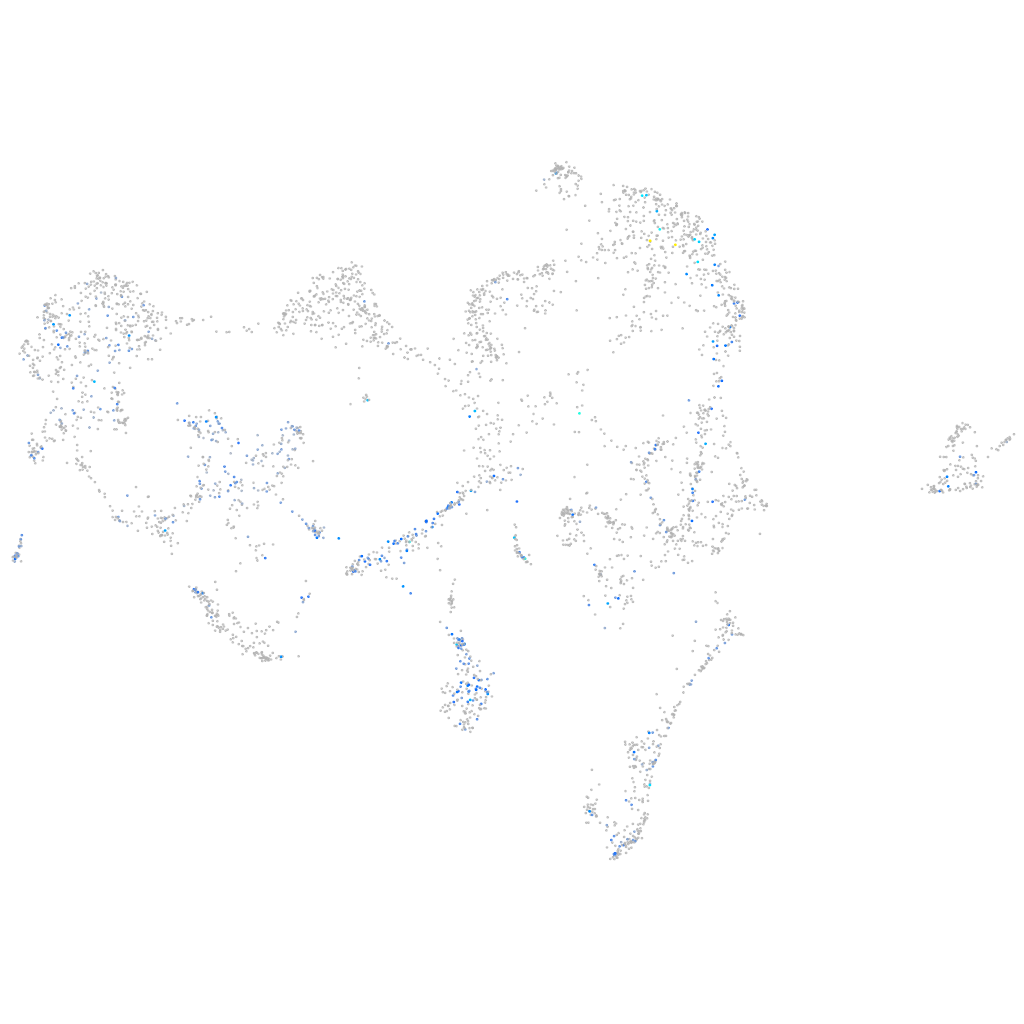
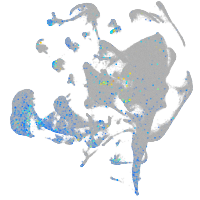
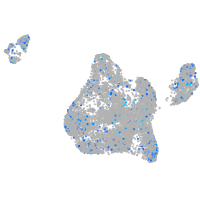
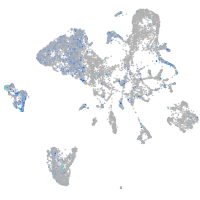
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ca15a | 0.223 | atp1a1a.2 | -0.101 |
| CDK18 | 0.217 | BX000438.2 | -0.098 |
| atp6v0a1a | 0.215 | malb | -0.089 |
| ceacam1 | 0.210 | si:dkey-189h5.6 | -0.086 |
| slc4a1b | 0.205 | krt91 | -0.085 |
| slc6a6b | 0.195 | pcdh11 | -0.083 |
| zgc:193726 | 0.195 | prlra | -0.081 |
| slc31a2 | 0.195 | elovl1b | -0.080 |
| atp6v1aa | 0.193 | ifi45 | -0.079 |
| atp6v1ba | 0.193 | gcshb | -0.072 |
| foxi3a | 0.191 | slc12a10.2 | -0.072 |
| tfe3b | 0.188 | ndrg1a | -0.072 |
| slc41a1 | 0.188 | hmgn2 | -0.070 |
| si:dkey-192d15.2 | 0.187 | dap1b | -0.069 |
| chp1 | 0.187 | ahcyl1 | -0.068 |
| clcn2b | 0.187 | slc4a4b | -0.068 |
| rpia | 0.183 | zgc:123295 | -0.067 |
| atp6v0ca | 0.181 | clcn2c | -0.066 |
| atp6ap1b | 0.178 | egr2a | -0.066 |
| cox5b2 | 0.176 | ckbb | -0.066 |
| atp6v1f | 0.175 | zgc:101785 | -0.065 |
| atp6v0d1 | 0.173 | trim35-12 | -0.062 |
| etv5a | 0.172 | gsto1 | -0.062 |
| etf1a | 0.170 | kcnj1a.2 | -0.061 |
| atp6v1g1 | 0.169 | fa2h | -0.061 |
| gcm2 | 0.169 | kcnj1a.6 | -0.061 |
| abca12 | 0.167 | pcdh10a | -0.061 |
| FAM135B | 0.167 | fam107b | -0.060 |
| degs1 | 0.165 | mef2aa | -0.060 |
| atp6ap2 | 0.165 | atp1a1a.3 | -0.060 |
| rnaseka | 0.162 | nfe2 | -0.059 |
| scgn | 0.161 | zgc:163083 | -0.058 |
| atp6v1c1b | 0.160 | kcnj1a.5 | -0.058 |
| atp6v1d | 0.160 | s100z | -0.056 |
| pfas | 0.158 | lgals3b | -0.056 |