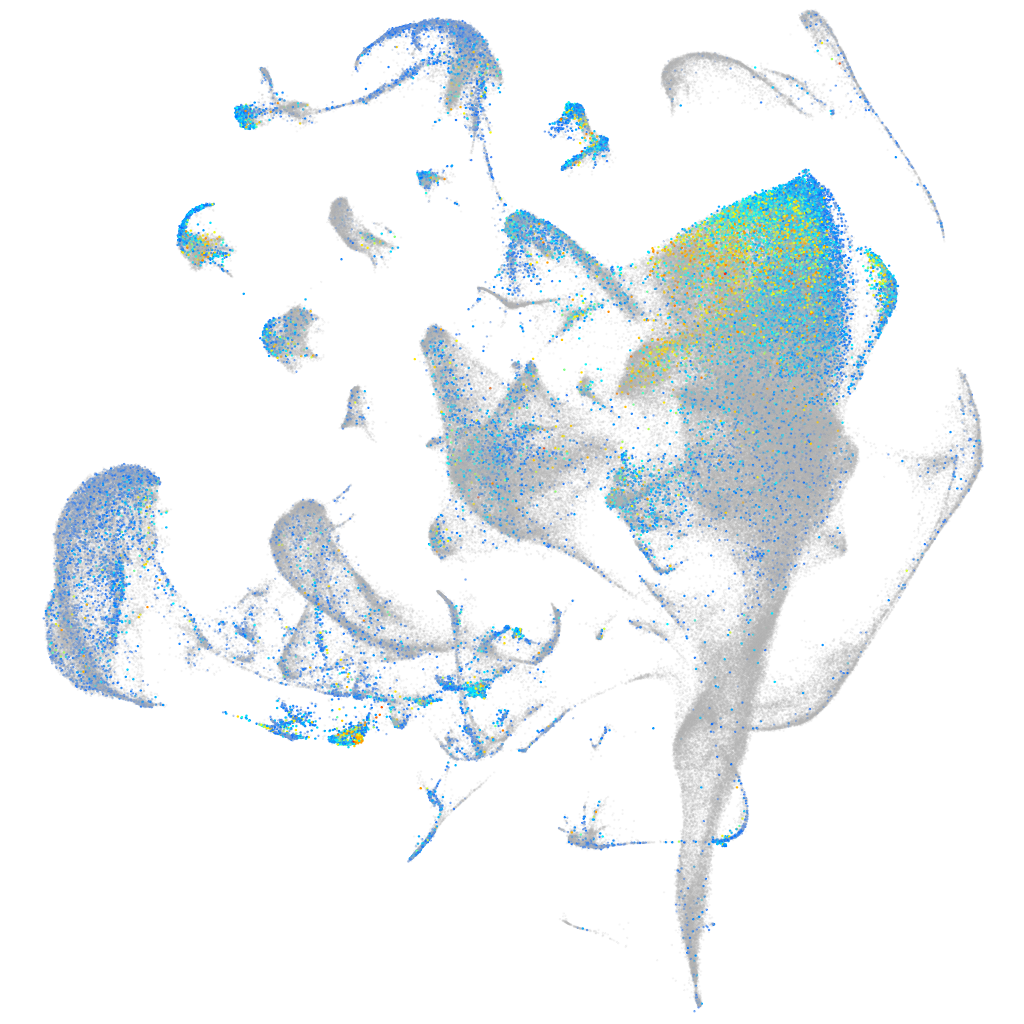ATPase H+ transporting V1 subunit C1b
ZFIN 
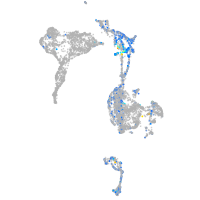
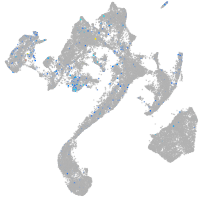
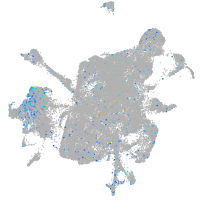
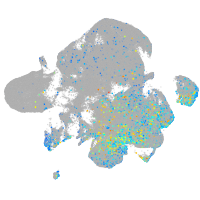
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.250 | hspb1 | -0.112 |
| atp6v1g1 | 0.245 | cx43.4 | -0.110 |
| atp6v1h | 0.213 | akap12b | -0.101 |
| atp6ap2 | 0.211 | dkc1 | -0.099 |
| zgc:65894 | 0.201 | lig1 | -0.099 |
| atp6v0d1 | 0.200 | nop58 | -0.098 |
| ywhag2 | 0.197 | fbl | -0.097 |
| sncb | 0.196 | mki67 | -0.096 |
| atp6v0cb | 0.190 | hnrnpa1b | -0.095 |
| gapdhs | 0.188 | id1 | -0.095 |
| stmn2a | 0.188 | pou5f3 | -0.095 |
| calm1a | 0.184 | chaf1a | -0.093 |
| gng3 | 0.181 | si:ch211-152c2.3 | -0.093 |
| vamp2 | 0.181 | nop56 | -0.091 |
| snap25a | 0.179 | stm | -0.091 |
| stxbp1a | 0.179 | polr3gla | -0.090 |
| syn2a | 0.176 | stmn1a | -0.090 |
| tpi1b | 0.176 | banf1 | -0.089 |
| calm1b | 0.175 | npm1a | -0.089 |
| rnasekb | 0.175 | si:dkey-66i24.9 | -0.089 |
| atp6v0b | 0.173 | ccnd1 | -0.088 |
| eno2 | 0.173 | zgc:110425 | -0.086 |
| ndrg3a | 0.171 | anp32b | -0.085 |
| atp1b1b | 0.170 | apoeb | -0.085 |
| pkma | 0.169 | tpx2 | -0.085 |
| syngr1a | 0.169 | asb11 | -0.084 |
| atpv0e2 | 0.168 | bms1 | -0.084 |
| ndufa4 | 0.168 | ccnb1 | -0.084 |
| atp6v1aa | 0.166 | gar1 | -0.084 |
| ldhba | 0.166 | ccna2 | -0.083 |
| mllt11 | 0.165 | ddx18 | -0.083 |
| sncgb | 0.165 | fbxo5 | -0.083 |
| stx1b | 0.165 | msna | -0.082 |
| map1aa | 0.164 | nasp | -0.082 |
| eno1a | 0.163 | tbx16 | -0.082 |