phosphoribosylformylglycinamidine synthase
ZFIN 
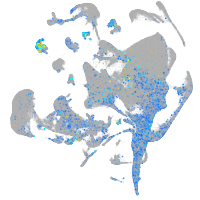
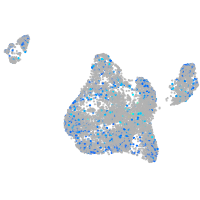
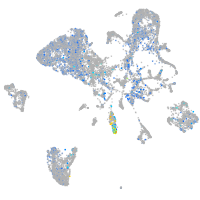
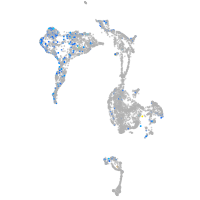
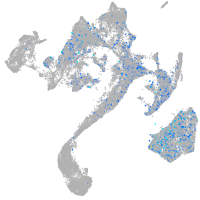
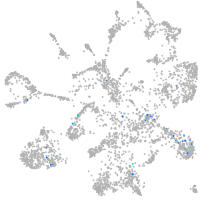
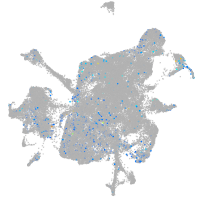
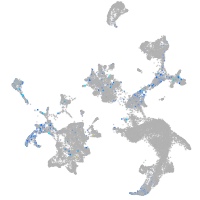
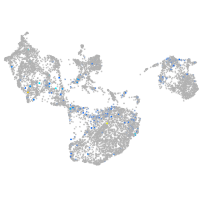
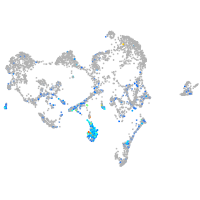
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc2a15a | 0.281 | gpm6aa | -0.098 |
| paics | 0.248 | rtn1a | -0.091 |
| aox5 | 0.233 | marcksl1a | -0.090 |
| pts | 0.229 | tuba1c | -0.089 |
| tmem130 | 0.229 | elavl3 | -0.088 |
| slc22a7a | 0.226 | stmn1b | -0.088 |
| si:dkey-251i10.2 | 0.222 | rnasekb | -0.081 |
| uraha | 0.222 | ckbb | -0.078 |
| atic | 0.218 | tmsb | -0.077 |
| gch2 | 0.217 | nova2 | -0.076 |
| slc2a11b | 0.213 | gng3 | -0.075 |
| bscl2l | 0.210 | myt1b | -0.070 |
| CABZ01021592.1 | 0.208 | sncb | -0.070 |
| si:dkey-21a6.5 | 0.206 | atp6v0cb | -0.069 |
| sprb | 0.200 | vamp2 | -0.067 |
| LOC103910009 | 0.194 | ccni | -0.066 |
| gart | 0.191 | gpm6ab | -0.066 |
| cax1 | 0.190 | ip6k2a | -0.066 |
| gmps | 0.189 | fez1 | -0.065 |
| akr1b1 | 0.188 | si:dkeyp-75h12.5 | -0.065 |
| ednrba | 0.186 | gng2 | -0.064 |
| oacyl | 0.186 | elavl4 | -0.063 |
| impdh1b | 0.183 | pvalb1 | -0.062 |
| prps1a | 0.179 | tnrc6c1 | -0.062 |
| rab34b | 0.178 | tubb5 | -0.062 |
| CABZ01048402.2 | 0.175 | wu:fb55g09 | -0.062 |
| ppat | 0.173 | calm1a | -0.062 |
| pnp4a | 0.172 | myt1a | -0.060 |
| si:ch211-256m1.8 | 0.171 | pvalb2 | -0.060 |
| unm-sa821 | 0.169 | stx1b | -0.060 |
| gpr143 | 0.168 | actc1b | -0.059 |
| si:ch211-108c6.2 | 0.168 | fam168a | -0.059 |
| phyhd1 | 0.167 | mllt11 | -0.059 |
| zgc:110789 | 0.167 | ywhag2 | -0.059 |
| gpnmb | 0.164 | csdc2a | -0.058 |