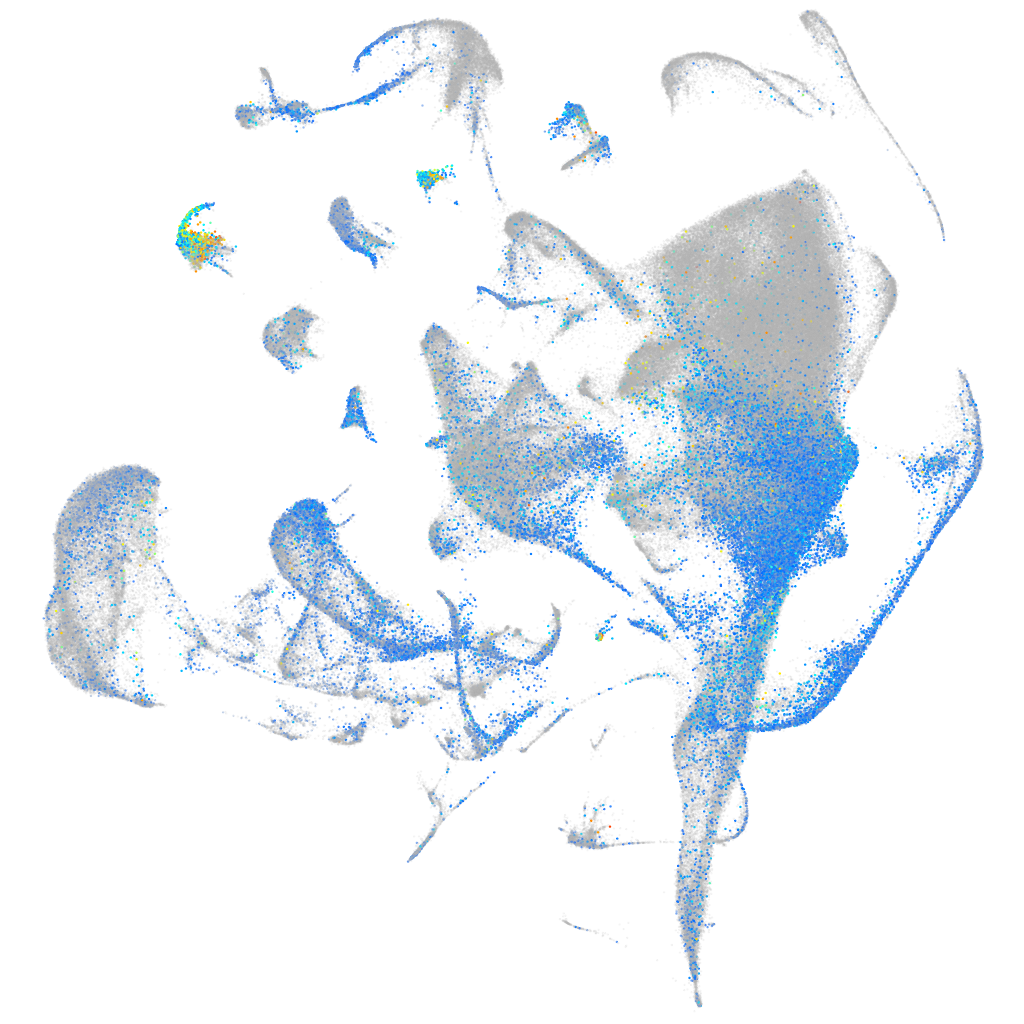phosphoribosylglycinamide formyltransferase
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paics | 0.311 | elavl3 | -0.102 |
| atic | 0.282 | stmn1b | -0.095 |
| slc2a15a | 0.244 | gpm6aa | -0.093 |
| prps1a | 0.231 | rtn1a | -0.090 |
| npm1a | 0.208 | tmsb | -0.089 |
| ppat | 0.206 | myt1b | -0.086 |
| pts | 0.204 | marcksl1a | -0.084 |
| aox5 | 0.203 | ckbb | -0.082 |
| impdh1b | 0.203 | myt1a | -0.082 |
| shmt1 | 0.201 | tuba1c | -0.081 |
| tmem130 | 0.197 | insm1a | -0.076 |
| sprb | 0.196 | scrt2 | -0.075 |
| gmps | 0.194 | nova2 | -0.071 |
| pfas | 0.191 | si:dkey-28b4.7 | -0.071 |
| snu13b | 0.190 | gpm6ab | -0.070 |
| slc22a7a | 0.189 | rnasekb | -0.070 |
| gch2 | 0.187 | tubb5 | -0.070 |
| slc2a11b | 0.185 | gng3 | -0.069 |
| uraha | 0.182 | ip6k2a | -0.069 |
| si:dkey-251i10.2 | 0.181 | tuba1a | -0.069 |
| cax1 | 0.177 | ebf3a | -0.069 |
| nop10 | 0.177 | pvalb1 | -0.068 |
| nhp2 | 0.176 | tp53inp2 | -0.068 |
| si:dkey-21a6.5 | 0.176 | hmgb3a | -0.067 |
| CABZ01021592.1 | 0.174 | pvalb2 | -0.067 |
| LOC103910009 | 0.173 | actc1b | -0.066 |
| nop58 | 0.173 | gng2 | -0.066 |
| akr1b1 | 0.172 | btg1 | -0.065 |
| pa2g4a | 0.172 | chd4a | -0.065 |
| pnp4a | 0.172 | CU634008.1 | -0.065 |
| cad | 0.169 | LOC100537384 | -0.065 |
| dkc1 | 0.169 | ccni | -0.064 |
| ednrba | 0.169 | dlb | -0.064 |
| bscl2l | 0.168 | pbx1a | -0.064 |
| gpr143 | 0.167 | LOC798783 | -0.063 |