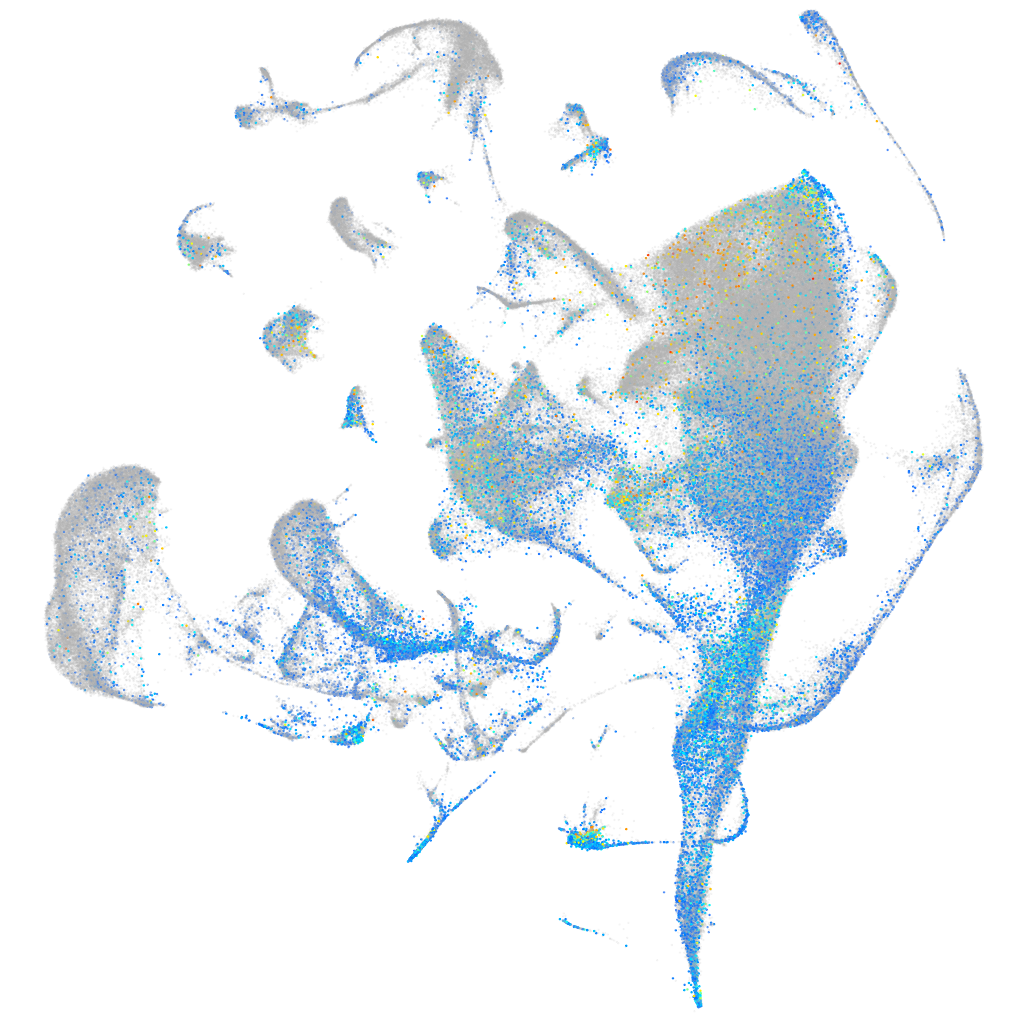ETS variant transcription factor 5a
ZFIN 
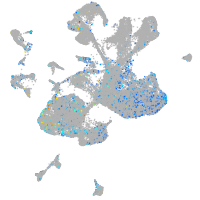
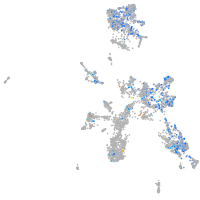
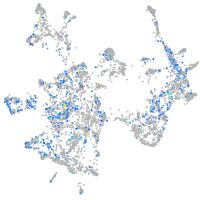
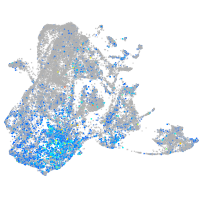
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cmn | 0.128 | rtn1a | -0.079 |
| zmp:0000000760 | 0.126 | stmn1b | -0.079 |
| tbxta | 0.124 | tuba1c | -0.079 |
| matn3b | 0.123 | gpm6ab | -0.077 |
| zgc:113232 | 0.123 | gpm6aa | -0.076 |
| si:dkey-234i14.3 | 0.122 | elavl3 | -0.073 |
| matn3a | 0.118 | hbbe1.3 | -0.071 |
| col9a1b | 0.117 | sncb | -0.071 |
| susd5 | 0.117 | hbae3 | -0.068 |
| chad | 0.116 | tmsb | -0.068 |
| id1 | 0.114 | atp6v0cb | -0.067 |
| hspb1 | 0.111 | ckbb | -0.066 |
| si:ch73-23l24.1 | 0.111 | gng3 | -0.066 |
| si:ch211-13f8.2 | 0.110 | hbae1.1 | -0.065 |
| si:ch211-106h4.6 | 0.109 | rtn1b | -0.065 |
| paqr4b | 0.108 | nova2 | -0.062 |
| nop58 | 0.106 | nsg2 | -0.060 |
| apoeb | 0.105 | rnasekb | -0.060 |
| akap12b | 0.104 | elavl4 | -0.057 |
| dkc1 | 0.104 | hbbe1.1 | -0.057 |
| etv5b | 0.104 | icn2 | -0.057 |
| zgc:175280 | 0.104 | pvalb1 | -0.057 |
| gas2b | 0.103 | pvalb2 | -0.057 |
| npm1a | 0.103 | vamp2 | -0.057 |
| epyc | 0.101 | fez1 | -0.056 |
| fbl | 0.101 | gng2 | -0.056 |
| igsf10 | 0.101 | myt1b | -0.056 |
| apela | 0.100 | tubb5 | -0.055 |
| cdx4 | 0.100 | csdc2a | -0.054 |
| chadla | 0.100 | gap43 | -0.054 |
| cx43.4 | 0.100 | tcnbb | -0.054 |
| nop56 | 0.100 | cnrip1a | -0.053 |
| col8a1a | 0.098 | si:dkeyp-75h12.5 | -0.053 |
| si:ch211-152c2.3 | 0.097 | stx1b | -0.053 |
| plod1a | 0.096 | krtt1c19e | -0.052 |