si:ch211-106h4.6
ZFIN 
Other cell groups


all cells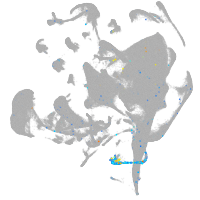 |
blastomeres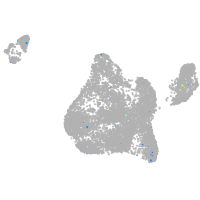 |
PGCs |
endoderm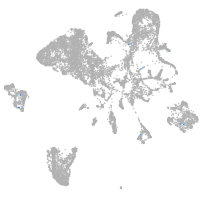 |
axial mesoderm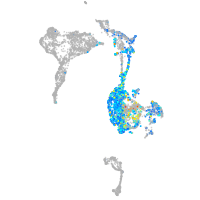 |
muscle  |
mural cells / non-skeletal muscle 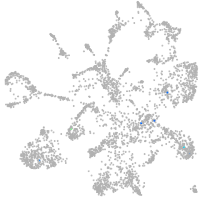 |
mesenchyme 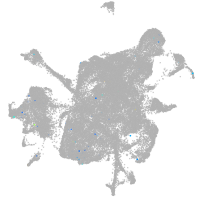 |
hematopoietic / vasculature 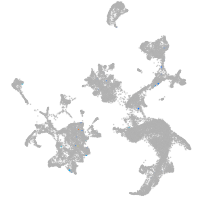 |
pronephros  |
neural |
spinal cord / glia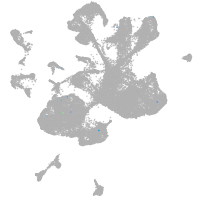 |
eye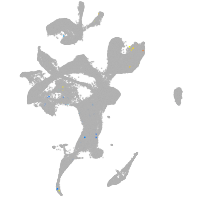 |
taste / olfactory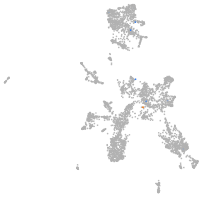 |
otic / lateral line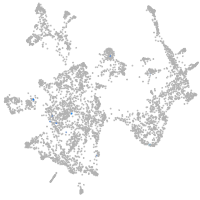 |
epidermis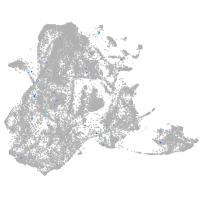 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cmn | 0.588 | marcksl1a | -0.042 |
| zmp:0000000760 | 0.587 | tuba1c | -0.041 |
| zgc:113232 | 0.582 | prdx2 | -0.040 |
| si:ch73-23l24.1 | 0.535 | nova2 | -0.039 |
| susd5 | 0.534 | jpt1b | -0.038 |
| si:dkey-234i14.3 | 0.527 | rtn1a | -0.037 |
| si:ch211-13f8.2 | 0.526 | tuba1a | -0.037 |
| paqr4b | 0.524 | fabp3 | -0.036 |
| chadla | 0.523 | gpm6aa | -0.036 |
| matn3b | 0.523 | hmgb1b | -0.036 |
| col9a1b | 0.511 | si:dkey-276j7.1 | -0.035 |
| chad | 0.490 | ckbb | -0.034 |
| matn3a | 0.477 | elavl3 | -0.034 |
| gas2b | 0.447 | marcksb | -0.034 |
| col8a1a | 0.437 | mdkb | -0.034 |
| igsf10 | 0.432 | si:ch211-137a8.4 | -0.034 |
| epyc | 0.410 | chd4a | -0.033 |
| LO018422.1 | 0.400 | hmgn3 | -0.033 |
| zgc:175280 | 0.400 | stmn1b | -0.032 |
| si:dkey-31g6.6 | 0.392 | gpm6ab | -0.031 |
| snorc | 0.390 | hmgb3a | -0.031 |
| si:dkey-1j5.4 | 0.383 | cct2 | -0.031 |
| col11a2 | 0.362 | mdh1aa | -0.030 |
| ucmab | 0.356 | rnasekb | -0.030 |
| cnmd | 0.349 | tmeff1b | -0.030 |
| tbx3b | 0.349 | zc4h2 | -0.030 |
| agtrap | 0.339 | hdac1 | -0.029 |
| col9a3 | 0.331 | hnrnpa0a | -0.029 |
| golim4b | 0.331 | ilf3b | -0.029 |
| si:dkey-174i8.1 | 0.330 | smarce1 | -0.029 |
| si:dkey-21p1.3 | 0.330 | ube2e1 | -0.029 |
| col9a2 | 0.329 | apex1 | -0.028 |
| ehd2b | 0.319 | tmsb | -0.028 |
| si:dkey-6n6.1 | 0.318 | gapdhs | -0.027 |
| mmp23bb | 0.315 | gnb1b | -0.027 |




