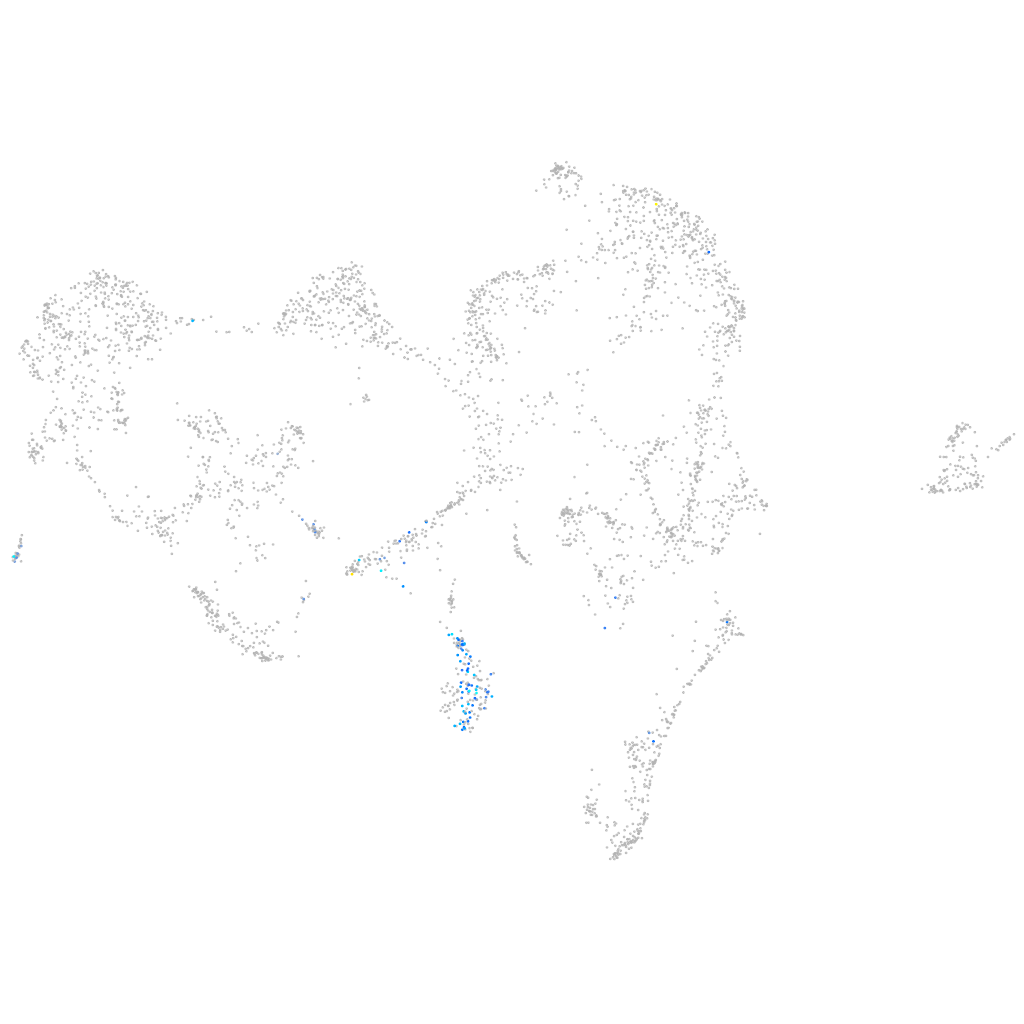
si:ch73-103l1.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ca15a | 0.382 | krt91 | -0.133 |
| ceacam1 | 0.362 | BX000438.2 | -0.125 |
| slc4a1b | 0.362 | aldob | -0.115 |
| atp6v0a1a | 0.353 | ucp2 | -0.114 |
| ca2 | 0.338 | ppiaa | -0.111 |
| pfas | 0.329 | fxyd6l | -0.105 |
| slc6a6a | 0.329 | ptmaa | -0.102 |
| atp6ap1b | 0.328 | calm2b | -0.102 |
| arhgap32a | 0.326 | rpl37 | -0.097 |
| scgn | 0.325 | adh5 | -0.096 |
| atp6v1ba | 0.307 | eif5a2 | -0.096 |
| atp6v1aa | 0.307 | ahcy | -0.095 |
| rpia | 0.305 | ptmab | -0.090 |
| slc31a2 | 0.303 | tcima | -0.090 |
| atp6v0ca | 0.302 | spaca4l | -0.088 |
| agrp | 0.301 | s100v1 | -0.086 |
| atp6v1g1 | 0.299 | rplp1 | -0.086 |
| si:ch211-147d7.5 | 0.297 | hmgn2 | -0.085 |
| atp6ap2 | 0.294 | cebpd | -0.084 |
| atp6v1c1b | 0.293 | calm2a | -0.084 |
| trpm7 | 0.291 | lmnb2 | -0.082 |
| atp6v1e1b | 0.290 | sdc4 | -0.081 |
| ptger1b | 0.289 | agr2 | -0.079 |
| arg2 | 0.288 | fa2h | -0.079 |
| foxi3a | 0.288 | si:ch211-222l21.1 | -0.079 |
| casq1b | 0.283 | atp1a1a.2 | -0.079 |
| gdi1 | 0.283 | tubb2b | -0.079 |
| atp6v1d | 0.282 | nap1l1 | -0.078 |
| rgs4 | 0.280 | slc38a5b | -0.078 |
| rnaseka | 0.280 | krt5 | -0.078 |
| etv5a | 0.278 | her6 | -0.078 |
| FAM135B | 0.277 | si:dkey-151g10.6 | -0.077 |
| atp6v0d1 | 0.276 | anp32b | -0.077 |
| clcn2b | 0.275 | tuba8l | -0.076 |
| gb:bc139872 | 0.275 | rpl27a | -0.076 |