si:ch211-180n24.3
ZFIN 
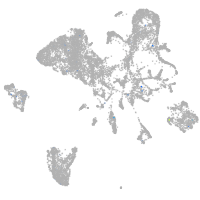
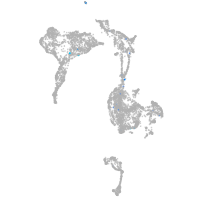
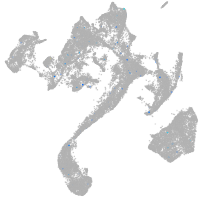
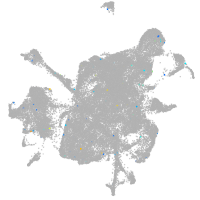
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| capn3a | 0.062 | elavl3 | -0.018 |
| BX548032.3 | 0.061 | stmn1b | -0.015 |
| cryba1l2 | 0.061 | rtn1a | -0.014 |
| crygm2d5 | 0.059 | gpm6aa | -0.013 |
| crygm2d12 | 0.055 | myt1a | -0.013 |
| lim2.5 | 0.055 | myt1b | -0.013 |
| bfsp2 | 0.054 | gng3 | -0.012 |
| crygm2d4 | 0.054 | rtn1b | -0.012 |
| lim2.4 | 0.053 | tmsb | -0.012 |
| cx23 | 0.052 | tuba1c | -0.012 |
| mipa | 0.052 | tubb5 | -0.012 |
| lim2.1 | 0.051 | chd4a | -0.011 |
| crybb1l3 | 0.050 | gpm6ab | -0.011 |
| lim2.3 | 0.050 | hmgb3a | -0.011 |
| crygm2d21 | 0.050 | scrt2 | -0.011 |
| cryba2a | 0.049 | si:dkey-276j7.1 | -0.011 |
| crygm2d20 | 0.049 | tuba1a | -0.011 |
| crygmxl2 | 0.049 | zc4h2 | -0.011 |
| mipb | 0.049 | csdc2a | -0.010 |
| endou2 | 0.049 | dlb | -0.010 |
| BFSP1 | 0.048 | elavl4 | -0.010 |
| crygm2d3 | 0.048 | epb41a | -0.010 |
| crygm2d10 | 0.048 | fez1 | -0.010 |
| crygm2d7 | 0.048 | gng2 | -0.010 |
| cryba1l1 | 0.047 | insm1a | -0.010 |
| crybb1l2 | 0.047 | jagn1a | -0.010 |
| crygm2d1 | 0.047 | nova2 | -0.010 |
| crygm2d18 | 0.047 | nsg2 | -0.010 |
| gja8b | 0.047 | sncb | -0.010 |
| crybb1l1 | 0.046 | tmeff1b | -0.010 |
| BX545855.1 | 0.046 | LOC100537384 | -0.010 |
| crygm2d13 | 0.046 | adam22 | -0.009 |
| crygm2d8 | 0.045 | aplp1 | -0.009 |
| gpx9 | 0.045 | atp6v0cb | -0.009 |
| cryba1b | 0.044 | ckbb | -0.009 |