sterile alpha motif domain containing 10b
ZFIN 
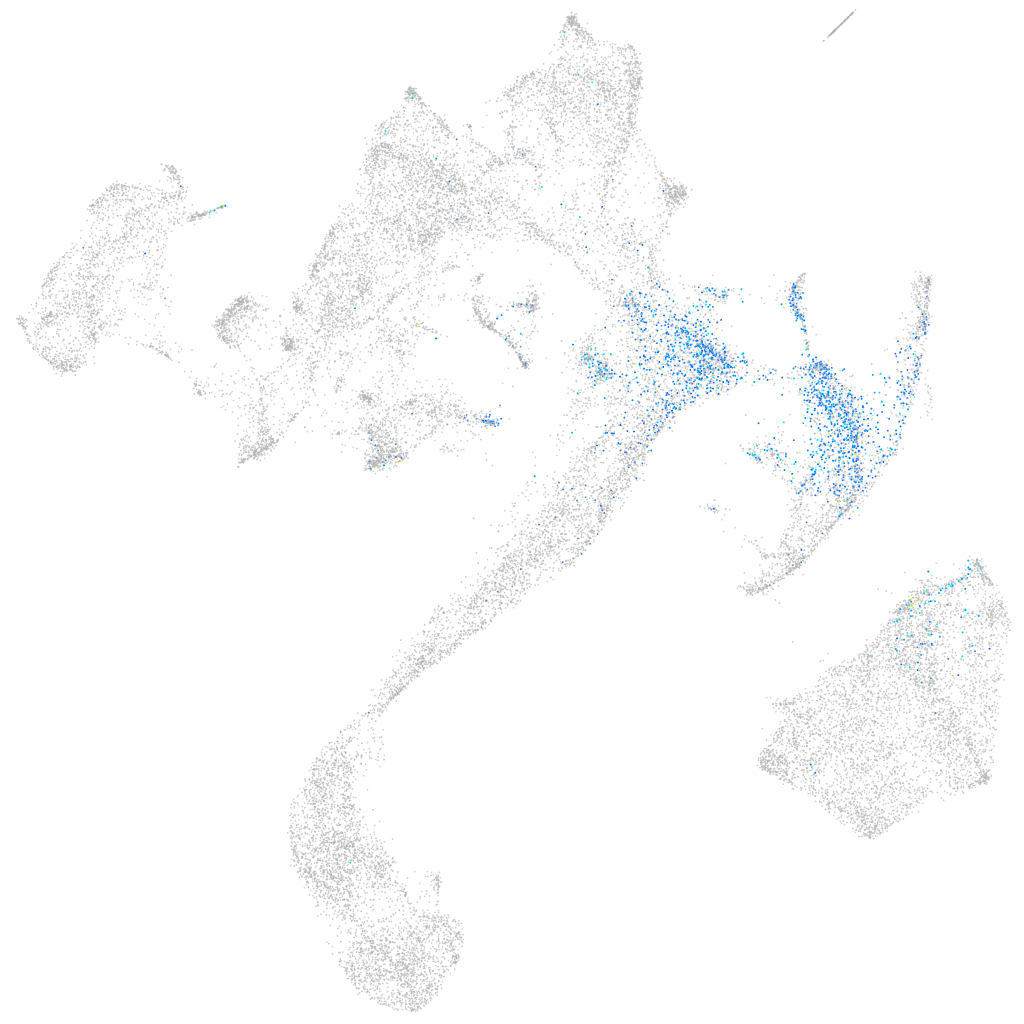
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fn1b | 0.327 | actc1b | -0.150 |
| meox1 | 0.318 | bhmt | -0.147 |
| draxin | 0.308 | fabp3 | -0.145 |
| pcdh8 | 0.296 | ckma | -0.143 |
| tbx6 | 0.283 | ckmb | -0.142 |
| tjp2b | 0.282 | atp2a1 | -0.141 |
| tcf15 | 0.276 | tmem38a | -0.137 |
| myf5 | 0.270 | si:dkey-16p21.8 | -0.136 |
| ripply1 | 0.269 | eno1a | -0.136 |
| BX001014.2 | 0.248 | ak1 | -0.135 |
| her1 | 0.247 | pabpc4 | -0.135 |
| apoc1 | 0.245 | gapdh | -0.134 |
| tuba8l2 | 0.240 | mylpfa | -0.133 |
| kazald2 | 0.238 | aldoab | -0.132 |
| her11 | 0.231 | tnnc2 | -0.127 |
| gnaia | 0.227 | acta1b | -0.125 |
| phc2a | 0.226 | neb | -0.124 |
| nid2a | 0.224 | pvalb1 | -0.123 |
| hoxb10a | 0.224 | srl | -0.123 |
| tspan7 | 0.218 | pmp22a | -0.122 |
| BX005254.3 | 0.217 | ldb3b | -0.122 |
| znfl1k | 0.215 | tpi1b | -0.122 |
| grin2da | 0.215 | pvalb2 | -0.121 |
| tma16 | 0.212 | mylz3 | -0.120 |
| ripply2 | 0.212 | myl1 | -0.120 |
| XLOC-042229 | 0.211 | cav3 | -0.119 |
| lmo4b | 0.209 | ndrg2 | -0.117 |
| igsf9a | 0.206 | col1a2 | -0.117 |
| dmrt2a | 0.202 | marcksl1b | -0.117 |
| hnrnpa0l | 0.195 | si:ch73-367p23.2 | -0.116 |
| XLOC-001964 | 0.193 | CABZ01078594.1 | -0.115 |
| hoxc3a | 0.193 | ldb3a | -0.115 |
| paics | 0.192 | gamt | -0.115 |
| emg1 | 0.191 | plecb | -0.115 |
| fgf17 | 0.191 | tnnt3a | -0.114 |