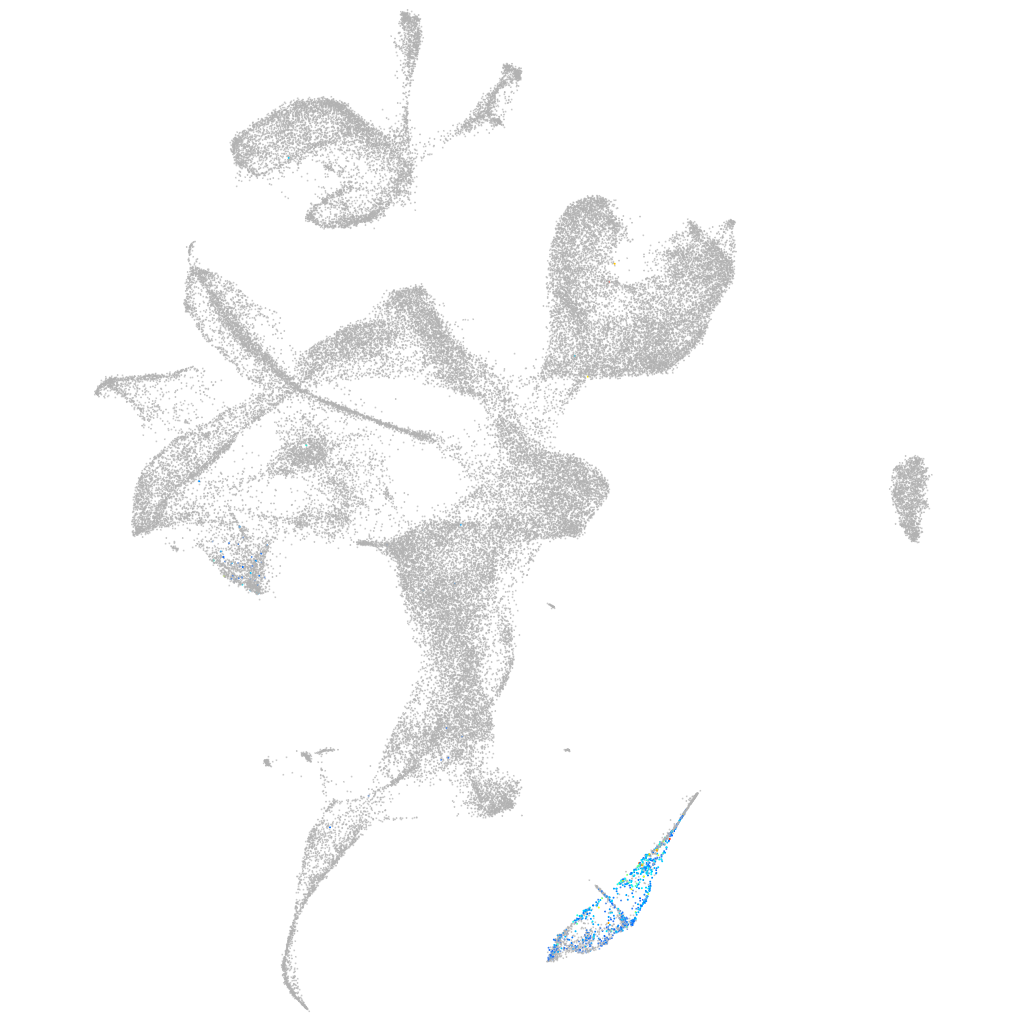
sphingosine-1-phosphate receptor 3a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 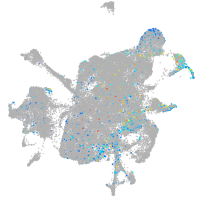 |
hematopoietic / vasculature 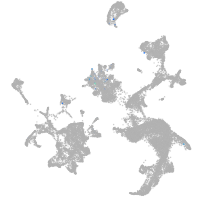 |
pronephros 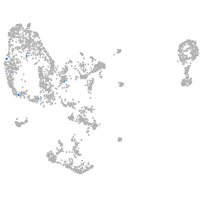 |
neural |
spinal cord / glia |
eye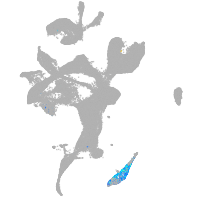 |
taste / olfactory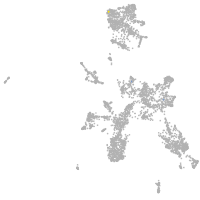 |
otic / lateral line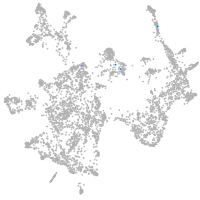 |
epidermis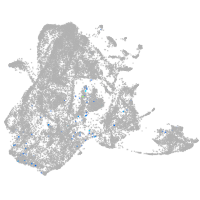 |
periderm |
fin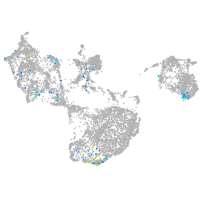 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gja8b | 0.609 | ptmab | -0.213 |
| otc | 0.547 | ptmaa | -0.151 |
| gja8a | 0.531 | hmgn6 | -0.143 |
| rbm24a | 0.522 | si:ch73-1a9.3 | -0.132 |
| pitx3 | 0.504 | hmgb1a | -0.123 |
| cps1 | 0.485 | tuba1c | -0.114 |
| prox2 | 0.467 | ckbb | -0.112 |
| CR356246.1 | 0.456 | h2afva | -0.108 |
| sparcl1 | 0.450 | tmsb4x | -0.105 |
| mipb | 0.447 | gpm6aa | -0.102 |
| prx | 0.447 | si:ch211-137a8.4 | -0.101 |
| cryba2a | 0.445 | fabp7a | -0.101 |
| si:ch211-214j24.14 | 0.444 | cadm3 | -0.096 |
| crybb1 | 0.440 | tuba1a | -0.094 |
| lim2.1 | 0.432 | hmgb1b | -0.093 |
| cryba1l1 | 0.430 | gpm6ab | -0.091 |
| cryba4 | 0.416 | h3f3c | -0.083 |
| cryba1b | 0.415 | anp32e | -0.083 |
| lim2.4 | 0.404 | hmgn2 | -0.083 |
| cryba2b | 0.403 | marcksb | -0.081 |
| cx23 | 0.402 | nova2 | -0.079 |
| lim2.3 | 0.394 | tubb2b | -0.078 |
| mipa | 0.392 | hmgb2a | -0.078 |
| crygn2 | 0.376 | mdkb | -0.076 |
| col28a1a | 0.373 | epb41a | -0.073 |
| crybb1l1 | 0.369 | ubc | -0.073 |
| si:dkeyp-44a8.4 | 0.367 | CR383676.1 | -0.072 |
| crybb1l2 | 0.364 | rorb | -0.069 |
| LOC110438076 | 0.348 | hnrnpa0a | -0.068 |
| crygmx | 0.347 | h3f3a | -0.067 |
| b3gnt5a | 0.344 | si:ch211-288g17.3 | -0.067 |
| mgarp | 0.340 | snap25b | -0.066 |
| slc7a11 | 0.339 | fabp3 | -0.066 |
| wnt7ba | 0.337 | cspg5a | -0.064 |
| foxe3 | 0.328 | stmn1b | -0.064 |