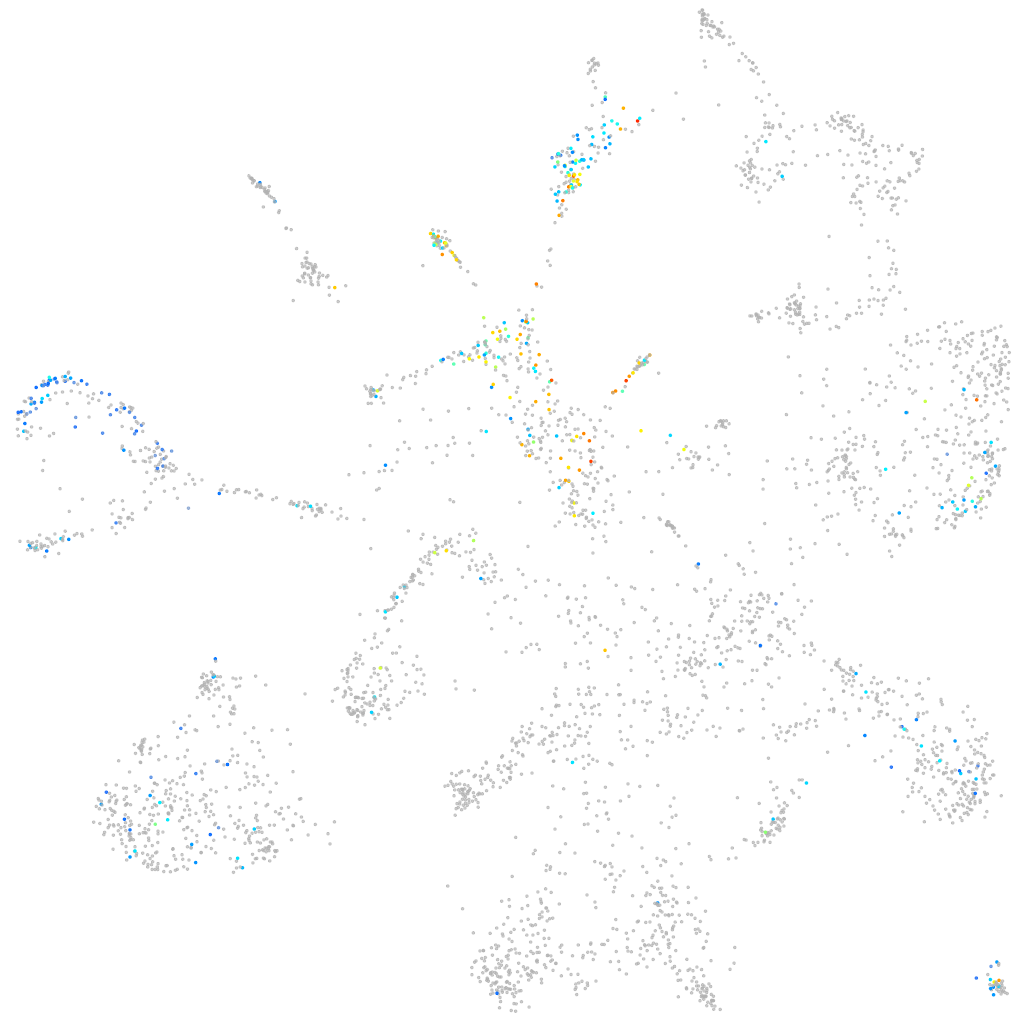
sphingosine-1-phosphate receptor 3a
ZFIN 
Other cell groups


all cells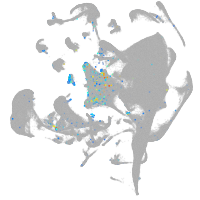 |
blastomeres |
PGCs |
endoderm |
axial mesoderm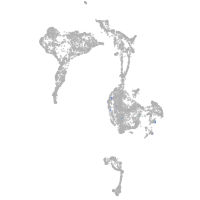 |
muscle 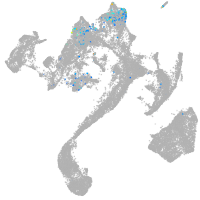 |
mural cells / non-skeletal muscle  |
mesenchyme 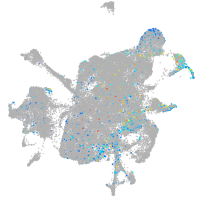 |
hematopoietic / vasculature 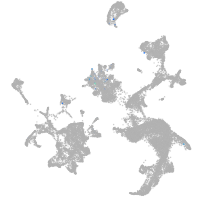 |
pronephros 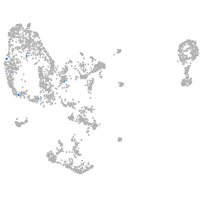 |
neural |
spinal cord / glia |
eye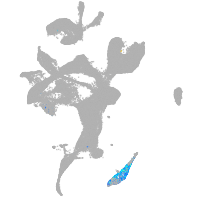 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-57k2.6 | 0.319 | nkx2.3 | -0.121 |
| rasl12 | 0.304 | hmga2 | -0.121 |
| fbln5 | 0.283 | si:dkeyp-66d1.7 | -0.116 |
| vim | 0.247 | hlx1 | -0.116 |
| loxa | 0.241 | desmb | -0.112 |
| htra1a | 0.239 | mylkb | -0.111 |
| crip1 | 0.236 | col6a2 | -0.109 |
| notch3 | 0.229 | si:ch211-62a1.3 | -0.108 |
| wfdc2 | 0.226 | ckbb | -0.108 |
| tpm4b | 0.220 | pdgfra | -0.106 |
| BX663503.3 | 0.216 | foxf1 | -0.106 |
| cd248a | 0.215 | foxp4 | -0.103 |
| cables2b | 0.212 | hmga1a | -0.102 |
| FP102018.1 | 0.210 | foxf2a | -0.101 |
| elnb | 0.207 | si:dkey-151g10.6 | -0.097 |
| hey2 | 0.203 | ntn5 | -0.096 |
| prss35 | 0.202 | cirbpa | -0.096 |
| si:dkey-164f24.2 | 0.202 | marcksl1a | -0.093 |
| itgb3b | 0.196 | rps28 | -0.092 |
| crip3 | 0.192 | si:dkey-56m19.5 | -0.091 |
| mcamb | 0.192 | col6a1 | -0.090 |
| fhl1b | 0.183 | snrpf | -0.090 |
| si:ch211-1a19.3 | 0.181 | ociad2 | -0.090 |
| gsc | 0.180 | XLOC-041870 | -0.089 |
| CR848683.2 | 0.179 | hmgb3a | -0.088 |
| pdlim2 | 0.177 | nkx3.3 | -0.088 |
| foxc1a | 0.177 | prdx1 | -0.086 |
| emid1 | 0.175 | col4a5 | -0.086 |
| ebf1b | 0.170 | sub1a | -0.086 |
| c7a | 0.169 | zgc:153704 | -0.085 |
| mef2cb | 0.169 | tmem88b | -0.084 |
| acana | 0.167 | rprmb | -0.084 |
| ebf1a | 0.166 | cald1b | -0.084 |
| hpgd | 0.166 | plpp1a | -0.083 |
| zeb2a | 0.166 | rpl34 | -0.083 |