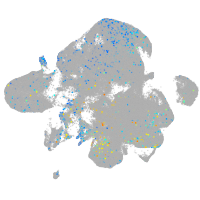
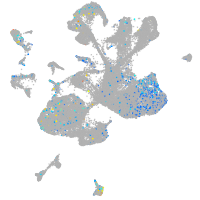
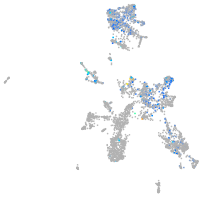
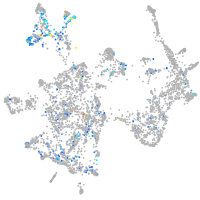
"retinoid X receptor, alpha a"
ZFIN 
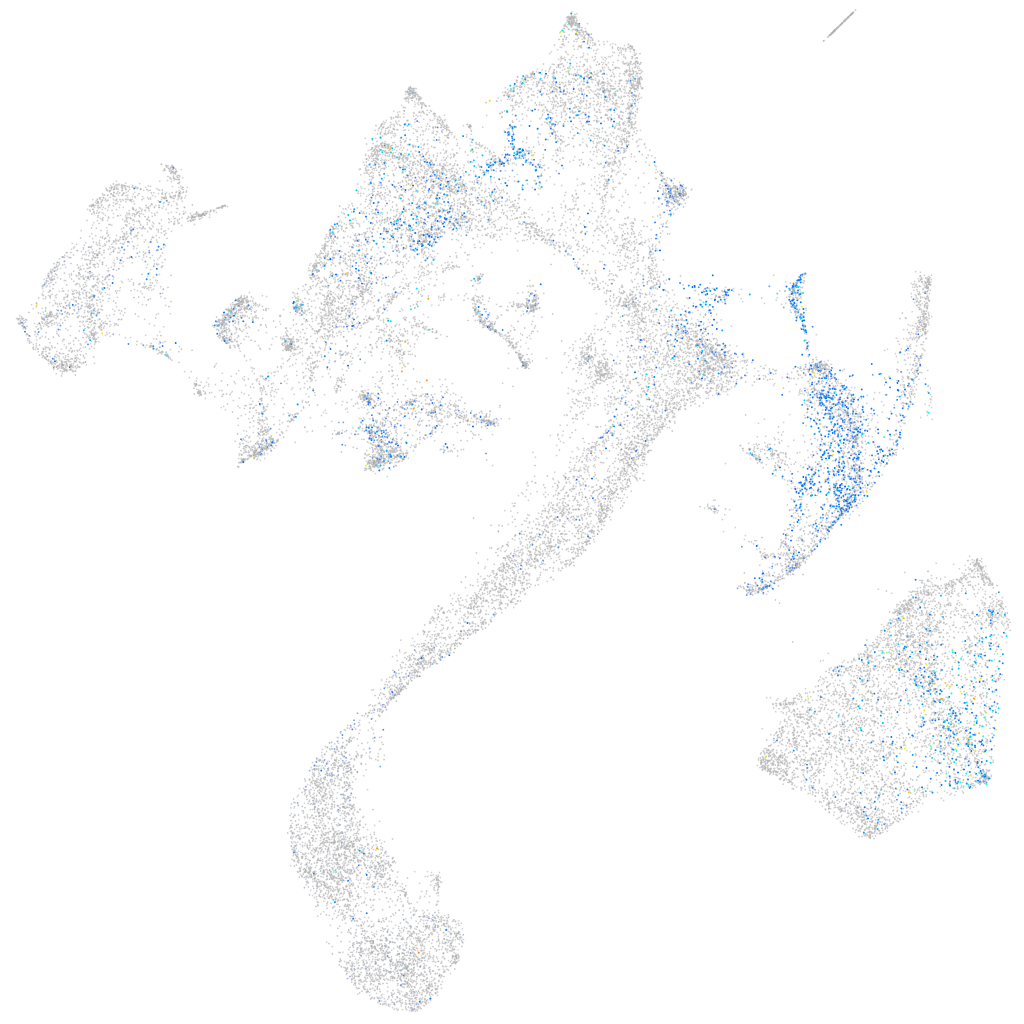
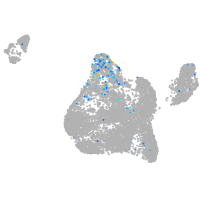
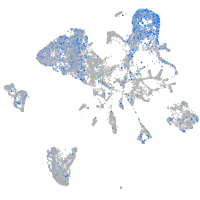
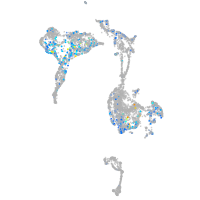
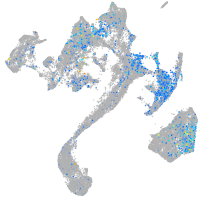
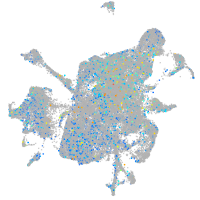
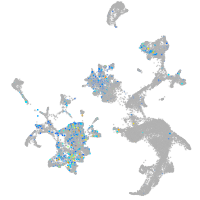
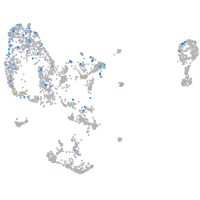
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoc1 | 0.205 | actc1b | -0.134 |
| msgn1 | 0.201 | ttn.2 | -0.126 |
| her1 | 0.183 | aldoab | -0.120 |
| her7 | 0.181 | tmem38a | -0.120 |
| efnb2b | 0.178 | klhl41b | -0.119 |
| tbx16l | 0.174 | ak1 | -0.118 |
| apoeb | 0.169 | mybphb | -0.116 |
| nid2a | 0.167 | acta1b | -0.115 |
| tbx16 | 0.167 | ckmb | -0.114 |
| fbln1 | 0.165 | pabpc4 | -0.114 |
| itm2cb | 0.162 | ttn.1 | -0.114 |
| hes6 | 0.160 | tnnc2 | -0.113 |
| pcdh8 | 0.160 | gamt | -0.113 |
| cx43.4 | 0.156 | eno1a | -0.112 |
| mespab | 0.149 | tpma | -0.112 |
| cxcl12b | 0.144 | ckma | -0.111 |
| rarga | 0.144 | ldb3a | -0.110 |
| aplnra | 0.144 | srl | -0.110 |
| hoxd12a | 0.144 | desma | -0.110 |
| cmtm6 | 0.141 | txlnbb | -0.108 |
| ism1 | 0.141 | atp2a1 | -0.108 |
| dlc | 0.141 | tnni2b.1 | -0.108 |
| hoxd10a | 0.140 | acta1a | -0.106 |
| lpar1 | 0.139 | zgc:92518 | -0.106 |
| gnaia | 0.137 | zgc:101853 | -0.105 |
| ddit4 | 0.136 | myl1 | -0.104 |
| CABZ01080702.1 | 0.135 | gatm | -0.104 |
| hoxb10a | 0.133 | CABZ01078594.1 | -0.104 |
| tbx6 | 0.133 | mylpfa | -0.103 |
| rbm38 | 0.133 | cav3 | -0.102 |
| ripply2 | 0.130 | neb | -0.101 |
| her12 | 0.129 | cox17 | -0.101 |
| hoxc13a | 0.129 | si:ch211-131k2.3 | -0.101 |
| ephb3 | 0.129 | gapdh | -0.100 |
| pnrc2 | 0.128 | rbfox1l | -0.099 |