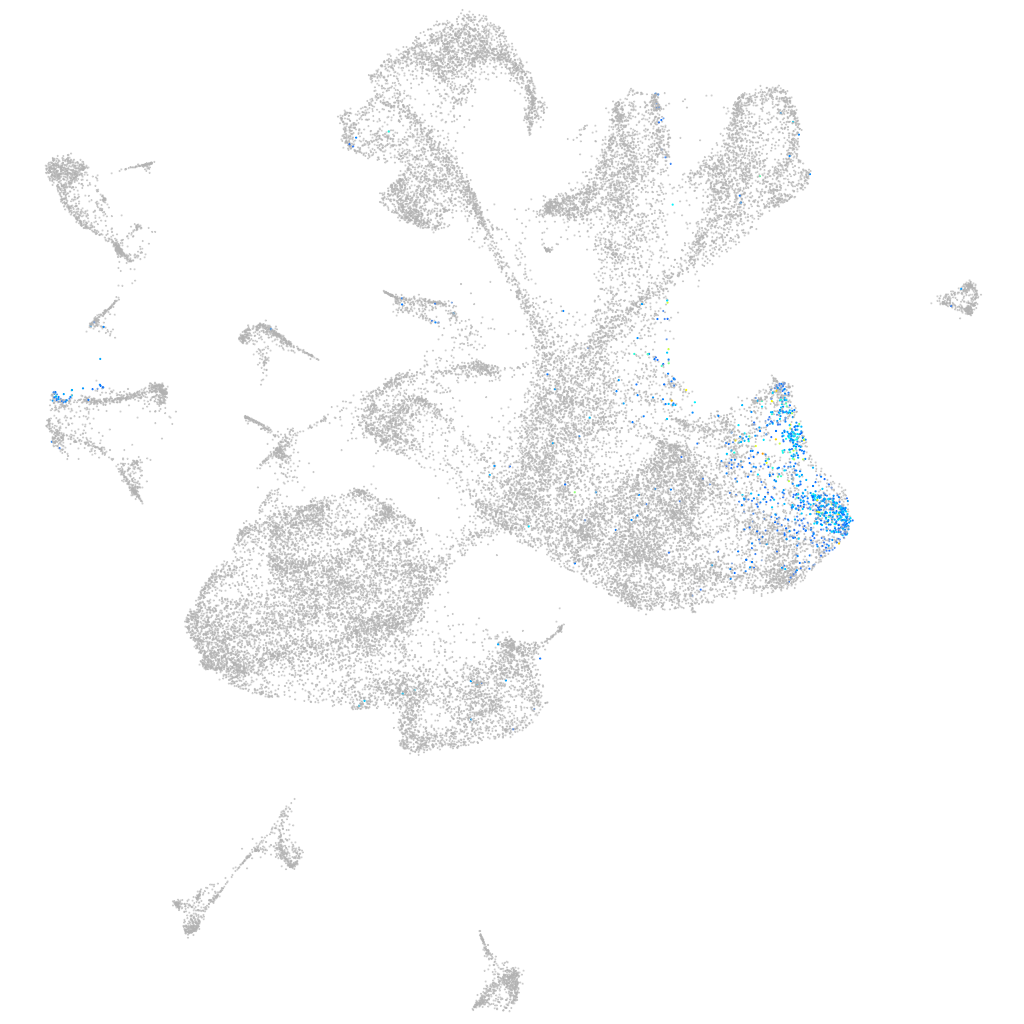
"POU domain, class 5, transcription factor 3"
ZFIN 
Other cell groups


all cells |
blastomeres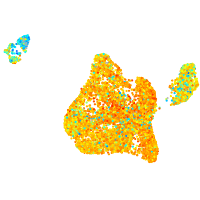 |
PGCs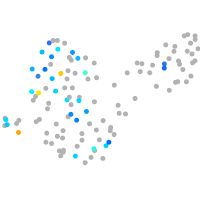 |
endoderm |
axial mesoderm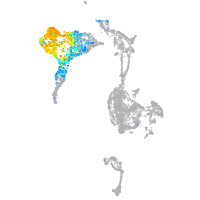 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural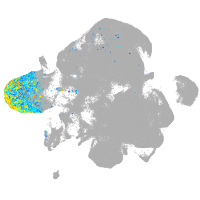 |
spinal cord / glia |
eye |
taste / olfactory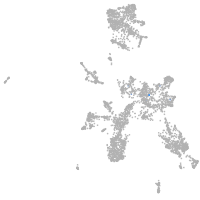 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous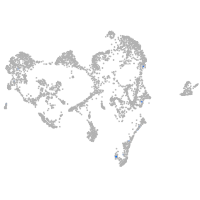 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sp5l | 0.437 | fabp3 | -0.169 |
| pnx | 0.401 | marcksl1b | -0.160 |
| cdx4 | 0.386 | gpm6aa | -0.143 |
| noto | 0.380 | tmsb4x | -0.139 |
| chrd | 0.368 | ckbb | -0.125 |
| nkx1.2la | 0.357 | CR383676.1 | -0.124 |
| foxd5 | 0.356 | fabp7a | -0.118 |
| dnase1l4.1 | 0.352 | rtn1a | -0.106 |
| fgf8a | 0.312 | meis1b | -0.106 |
| cxcr4a | 0.312 | CU467822.1 | -0.105 |
| nradd | 0.305 | nova2 | -0.103 |
| apela | 0.286 | pvalb1 | -0.100 |
| si:dkey-18j18.3 | 0.266 | zgc:165461 | -0.098 |
| apoc1 | 0.266 | pvalb2 | -0.097 |
| XLOC-042222 | 0.264 | tmsb | -0.090 |
| hspb1 | 0.263 | gpm6ab | -0.089 |
| BX005254.3 | 0.253 | nr2f1a | -0.089 |
| sall4 | 0.246 | CU634008.1 | -0.088 |
| hoxd12a | 0.243 | atp1a1b | -0.088 |
| si:dkeyp-110a12.4 | 0.243 | gpm6bb | -0.088 |
| hoxa13a | 0.243 | actc1b | -0.087 |
| znfl2a | 0.240 | stmn1b | -0.087 |
| eve1 | 0.239 | COX3 | -0.086 |
| lrrc17 | 0.235 | hbbe1.3 | -0.086 |
| aebp1 | 0.234 | slc1a2b | -0.083 |
| mllt3 | 0.230 | cspg5a | -0.082 |
| hoxd10a | 0.229 | rnasekb | -0.082 |
| spag6 | 0.225 | hbae3 | -0.081 |
| smkr1 | 0.223 | ppdpfb | -0.081 |
| cdx1a | 0.219 | si:dkeyp-75h12.5 | -0.080 |
| arid3b | 0.216 | si:ch211-133n4.4 | -0.079 |
| greb1 | 0.213 | atp1b4 | -0.078 |
| hoxa13b | 0.208 | si:dkey-276j7.1 | -0.078 |
| hoxb10a | 0.207 | si:ch73-386h18.1 | -0.078 |
| dpcd | 0.206 | pou3f3b | -0.077 |