myosin binding protein Ca
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 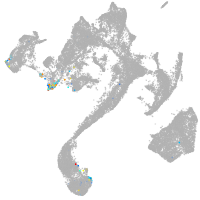 |
mural cells / non-skeletal muscle 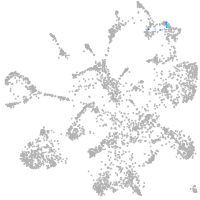 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 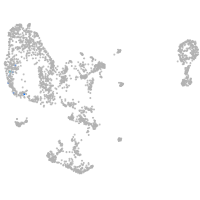 |
neural |
spinal cord / glia |
eye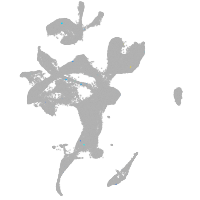 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni2a.3 | 0.161 | mdkb | -0.015 |
| tnni2a.1 | 0.152 | marcksl1a | -0.015 |
| mybpc1 | 0.144 | chd4a | -0.014 |
| zgc:172270 | 0.142 | nova2 | -0.014 |
| mybpha | 0.131 | si:ch211-288g17.3 | -0.013 |
| myoz3a | 0.124 | ddx39ab | -0.012 |
| obscna | 0.117 | mdka | -0.012 |
| mustn1b | 0.116 | si:ch211-137a8.4 | -0.012 |
| pvalb7 | 0.116 | si:dkey-56m19.5 | -0.012 |
| myhb | 0.114 | cbx3a | -0.011 |
| hspb11 | 0.106 | cspg5a | -0.011 |
| tnni2a.2 | 0.104 | CU467822.1 | -0.011 |
| si:rp71-17i16.4 | 0.098 | gpm6aa | -0.011 |
| CU681836.1 | 0.097 | h2afva | -0.011 |
| ryr1a | 0.097 | hdac1 | -0.011 |
| flnca | 0.096 | hmgb1b | -0.011 |
| tecra | 0.096 | hmgn3 | -0.011 |
| pvalb4 | 0.095 | hnrnpa0a | -0.011 |
| sync | 0.093 | hnrnph1l | -0.011 |
| si:dkey-211f22.5 | 0.091 | marcksb | -0.011 |
| fhl5 | 0.090 | rbbp4 | -0.011 |
| myoz1a | 0.090 | seta | -0.011 |
| mylk4a | 0.090 | setb | -0.011 |
| ankrd24 | 0.089 | XLOC-003690 | -0.011 |
| cavin1a | 0.089 | atrx | -0.010 |
| actn3b | 0.088 | gfap | -0.010 |
| AGBL1 | 0.088 | her12 | -0.010 |
| cav3 | 0.088 | ilf3b | -0.010 |
| cavin4a | 0.087 | nono | -0.010 |
| camk2a | 0.086 | nucks1a | -0.010 |
| klhl41a | 0.086 | pou3f1 | -0.010 |
| lmod2b | 0.086 | si:ch73-21g5.7 | -0.010 |
| casq2 | 0.085 | smarce1 | -0.010 |
| smpx | 0.085 | snrpb | -0.010 |
| tcap | 0.085 | snrpd1 | -0.010 |




