"trans-2,3-enoyl-CoA reductase a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm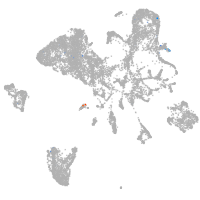 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mybpha | 0.159 | gpm6aa | -0.024 |
| casq1b | 0.151 | hmgb1b | -0.024 |
| zgc:172270 | 0.150 | si:ch211-288g17.3 | -0.024 |
| si:dkey-211f22.5 | 0.149 | marcksb | -0.023 |
| dhrs7cb | 0.142 | chd4a | -0.022 |
| jph2 | 0.138 | gpm6ab | -0.022 |
| CU681836.1 | 0.135 | nova2 | -0.022 |
| cavin4a | 0.133 | tuba1c | -0.022 |
| myoz1a | 0.132 | ckbb | -0.021 |
| tnni2a.3 | 0.132 | h2afva | -0.021 |
| actn3b | 0.130 | nucks1a | -0.021 |
| atp2a1l | 0.130 | tubb5 | -0.021 |
| casq2 | 0.130 | ywhaqb | -0.021 |
| hhatla | 0.129 | acin1a | -0.020 |
| si:ch211-266g18.10 | 0.127 | atrx | -0.020 |
| casq1a | 0.126 | gnb1b | -0.020 |
| ldb3b | 0.126 | hdac1 | -0.020 |
| myom1a | 0.125 | hnrnpa0a | -0.020 |
| hspb3 | 0.125 | hp1bp3 | -0.020 |
| xirp2b | 0.123 | mdkb | -0.020 |
| neb | 0.122 | rnasekb | -0.020 |
| si:ch73-367p23.2 | 0.122 | setb | -0.020 |
| cav3 | 0.121 | smarce1 | -0.020 |
| stac3 | 0.121 | tmeff1b | -0.020 |
| ckmt2b | 0.120 | marcksl1a | -0.020 |
| prr33 | 0.120 | elavl3 | -0.019 |
| mylk4a | 0.118 | si:dkey-276j7.1 | -0.019 |
| zgc:158296 | 0.118 | si:dkey-56m19.5 | -0.019 |
| actn3a | 0.117 | zc4h2 | -0.019 |
| ampd1 | 0.117 | alyref | -0.018 |
| nrap | 0.117 | calm3b | -0.018 |
| si:ch211-255p10.3 | 0.117 | cbx3a | -0.018 |
| pgam2 | 0.117 | cspg5a | -0.018 |
| si:ch211-157b11.14 | 0.117 | hmgn3 | -0.018 |
| cavin4b | 0.116 | hnrnpa1a | -0.018 |




