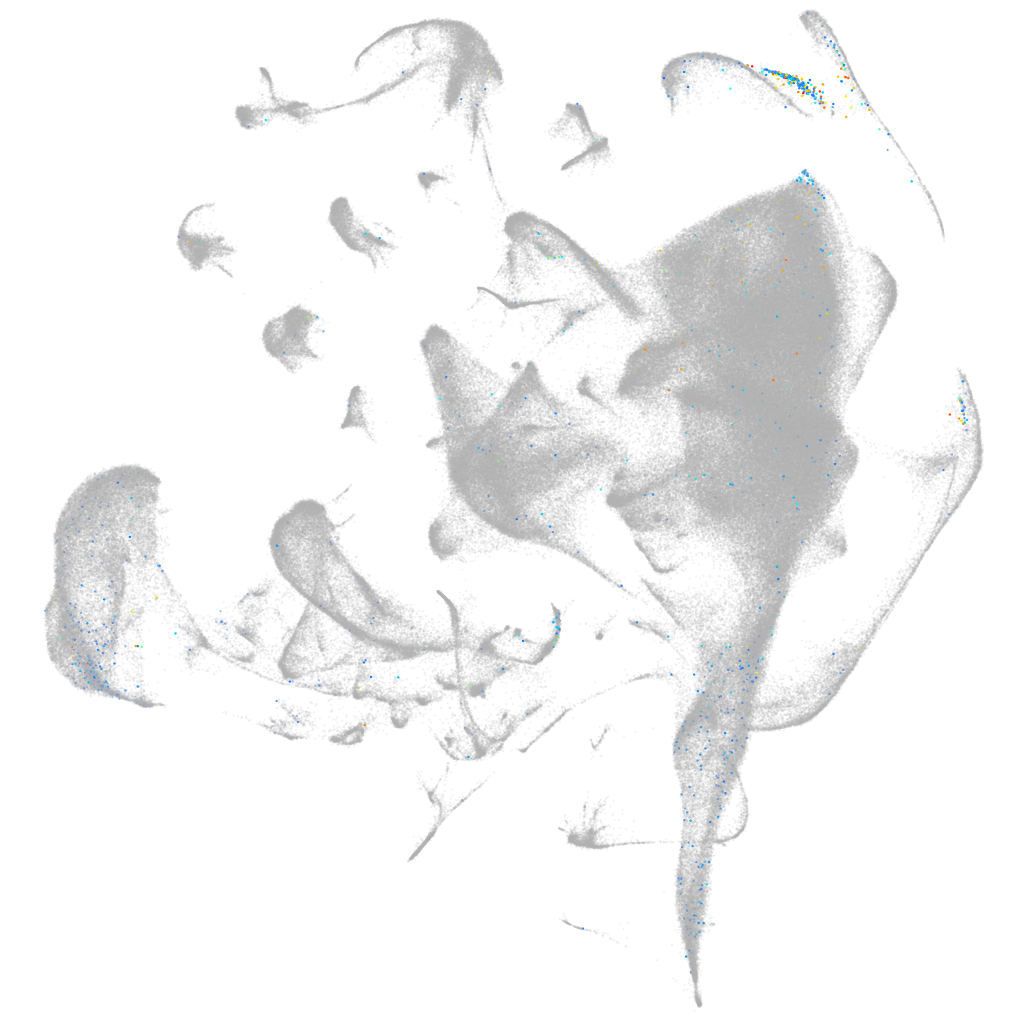"troponin I type 2a (skeletal, fast), tandem duplicate 2"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm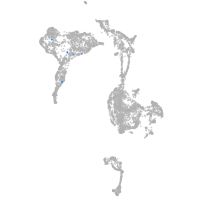 |
muscle 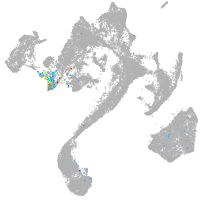 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 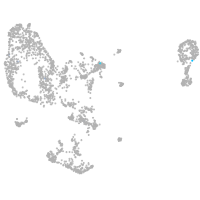 |
neural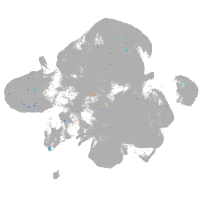 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni2a.3 | 0.218 | marcksl1a | -0.030 |
| tnni2a.1 | 0.213 | gpm6aa | -0.028 |
| myhb | 0.171 | gpm6ab | -0.027 |
| fhl5 | 0.161 | ckbb | -0.025 |
| zgc:172270 | 0.149 | rnasekb | -0.024 |
| mybpha | 0.147 | stmn1b | -0.024 |
| obscna | 0.147 | atp6v0cb | -0.023 |
| mybpc1 | 0.145 | cspg5a | -0.023 |
| ankrd24 | 0.130 | mdkb | -0.023 |
| sync | 0.130 | elavl3 | -0.022 |
| si:rp71-17i16.4 | 0.130 | hmgb1b | -0.022 |
| mustn1b | 0.122 | nova2 | -0.022 |
| pvalb7 | 0.121 | tuba1c | -0.021 |
| klhl41a | 0.119 | epb41a | -0.020 |
| cavin4a | 0.115 | gapdhs | -0.020 |
| AGBL1 | 0.114 | si:dkey-276j7.1 | -0.020 |
| cav3 | 0.113 | sncb | -0.020 |
| lmod2b | 0.113 | tmeff1b | -0.020 |
| tnni2b.1 | 0.113 | zc4h2 | -0.020 |
| myl4 | 0.111 | atp6v1e1b | -0.019 |
| myoz3a | 0.111 | fabp3 | -0.019 |
| myoz1a | 0.108 | rtn1b | -0.019 |
| pdlim3b | 0.107 | si:dkeyp-75h12.5 | -0.019 |
| casq2 | 0.106 | vamp2 | -0.019 |
| CU681836.1 | 0.106 | atpv0e2 | -0.018 |
| ryr1a | 0.106 | cadm3 | -0.018 |
| smpdl3b | 0.106 | tubb5 | -0.018 |
| cavin1a | 0.105 | calm1a | -0.018 |
| fhl2a | 0.104 | atp6ap2 | -0.017 |
| mybpc2a | 0.104 | fam168a | -0.017 |
| camk2a | 0.102 | fez1 | -0.017 |
| desma | 0.101 | gap43 | -0.017 |
| fhl1a | 0.100 | gnao1a | -0.017 |
| flnca | 0.098 | gng3 | -0.017 |
| si:dkey-211f22.5 | 0.097 | hbae3 | -0.017 |