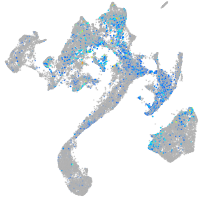
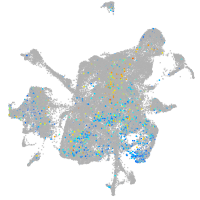
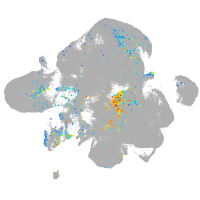
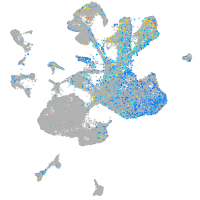
homeobox B6a
ZFIN 
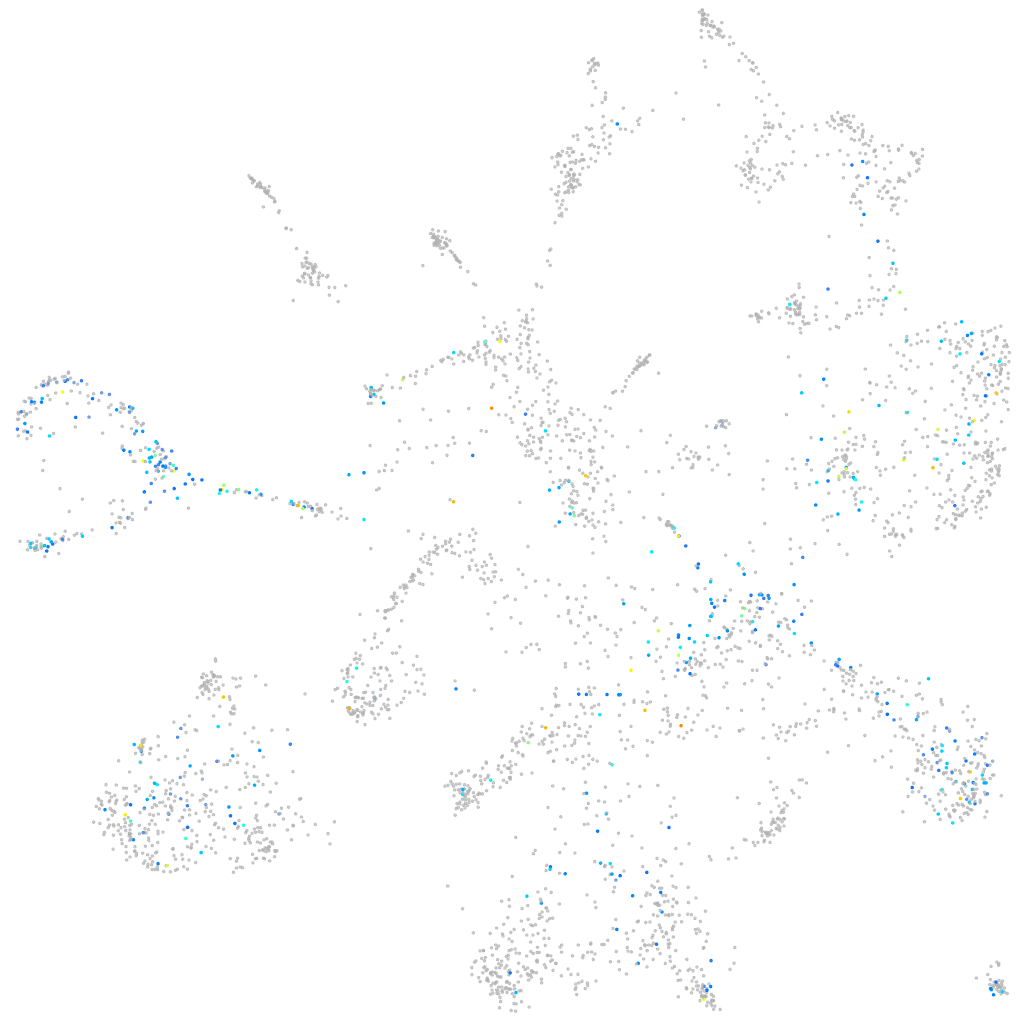
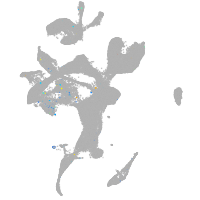
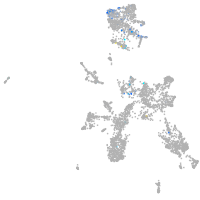
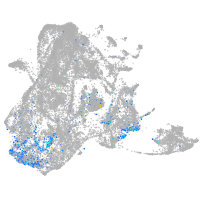
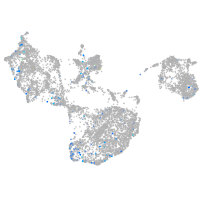
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxb5a | 0.298 | tmsb4x | -0.093 |
| hoxb5b | 0.233 | pcolcea | -0.069 |
| hoxb8a | 0.204 | cebpd | -0.069 |
| hoxc6a | 0.187 | hoxa10b | -0.065 |
| ldha | 0.181 | krt94 | -0.065 |
| hoxc3a | 0.171 | rgs2 | -0.064 |
| loxl5b | 0.168 | ednraa | -0.064 |
| pgk1 | 0.158 | mmel1 | -0.063 |
| wnt11r | 0.157 | ifitm1 | -0.062 |
| pkma | 0.156 | mfap5 | -0.062 |
| aldocb | 0.153 | dap1b | -0.060 |
| pgam1a | 0.147 | mcl1b | -0.060 |
| pitx1 | 0.146 | rbp4 | -0.060 |
| meis3 | 0.143 | lbh | -0.059 |
| hoxc8a | 0.142 | junba | -0.058 |
| eno3 | 0.139 | CELA1 (1 of many) | -0.058 |
| namptb | 0.136 | vim | -0.057 |
| hoxb6b | 0.135 | prss1 | -0.057 |
| gpia | 0.133 | hoxa11a | -0.057 |
| dtnbp1b | 0.131 | ntn1a | -0.056 |
| hoxc5a | 0.131 | ppdpfb | -0.056 |
| eno1a | 0.130 | pvalb2 | -0.056 |
| prdx1 | 0.129 | btg2 | -0.055 |
| hoxc1a | 0.126 | pvalb1 | -0.055 |
| inhbaa | 0.123 | socs3a | -0.055 |
| fbp2 | 0.122 | desmb | -0.054 |
| gapdhs | 0.122 | twist1b | -0.054 |
| VSTM2B | 0.122 | FP102018.1 | -0.053 |
| pfkpa | 0.121 | gata5 | -0.053 |
| alcama | 0.120 | si:ch211-137i24.10 | -0.053 |
| AL953907.1 | 0.120 | si:dkey-57k2.6 | -0.053 |
| barx1 | 0.119 | fhl1b | -0.052 |
| XLOC-008601 | 0.118 | ciarta | -0.052 |
| vasnb | 0.116 | c1qtnf5 | -0.052 |
| si:dkey-23a13.7 | 0.116 | rplp2 | -0.051 |