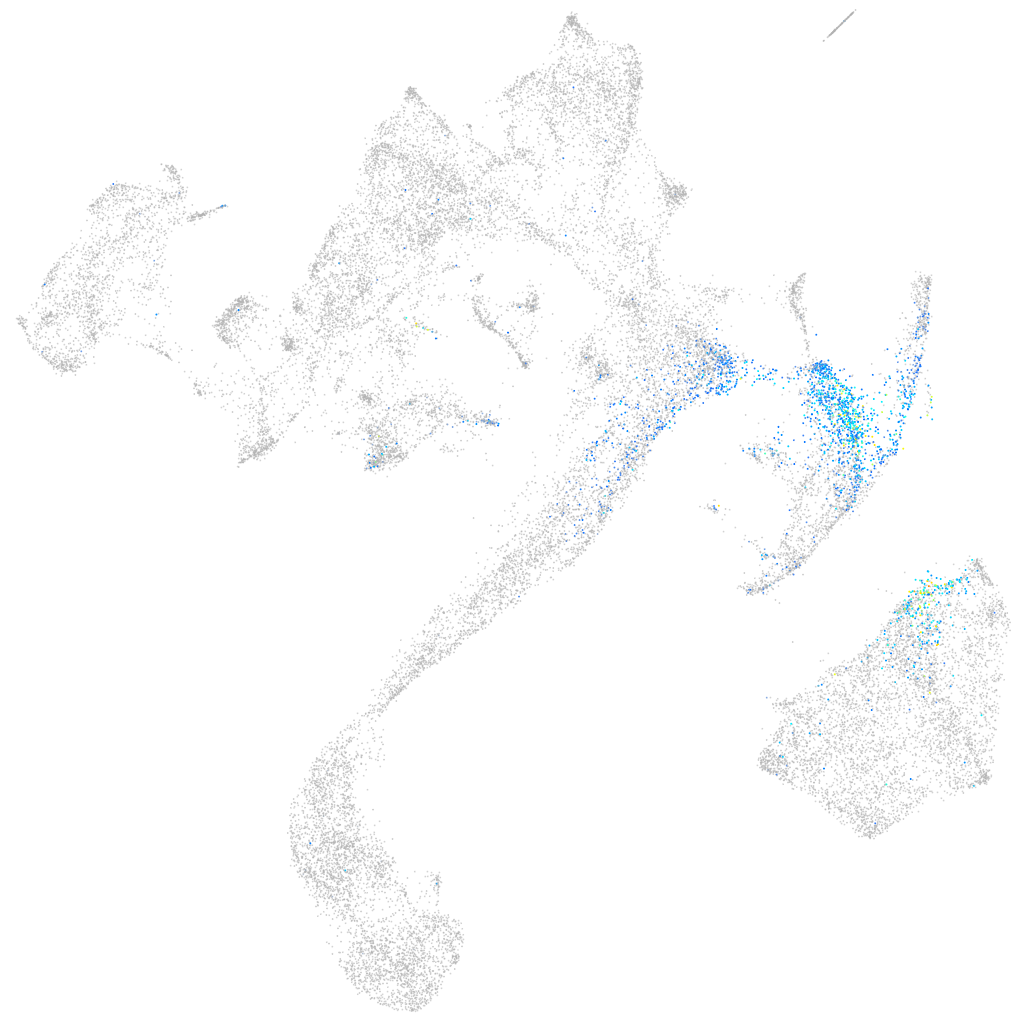
"glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tbx6 | 0.434 | fabp3 | -0.191 |
| pcdh8 | 0.371 | actc1b | -0.154 |
| myf5 | 0.350 | sparc | -0.153 |
| BX001014.2 | 0.337 | bhmt | -0.151 |
| draxin | 0.329 | ckma | -0.143 |
| fn1b | 0.325 | ak1 | -0.142 |
| her1 | 0.313 | ckmb | -0.141 |
| her11 | 0.308 | atp2a1 | -0.141 |
| ripply2 | 0.297 | eno1a | -0.137 |
| apoc1 | 0.284 | pabpc4 | -0.137 |
| BX005254.3 | 0.280 | eef1da | -0.136 |
| gnaia | 0.270 | mylpfa | -0.135 |
| meox1 | 0.261 | tmem38a | -0.135 |
| large2 | 0.258 | gapdh | -0.135 |
| hoxb7a | 0.258 | col1a2 | -0.130 |
| cpa6 | 0.253 | si:dkey-16p21.8 | -0.129 |
| XLOC-032526 | 0.252 | pmp22a | -0.129 |
| dlc | 0.251 | aldoab | -0.128 |
| mespaa | 0.250 | tpi1b | -0.125 |
| msgn1 | 0.245 | ndrg2 | -0.125 |
| tspan7 | 0.243 | acta1b | -0.125 |
| phc2a | 0.242 | eno3 | -0.125 |
| alpi.1 | 0.240 | tnnc2 | -0.124 |
| lmo4b | 0.238 | pvalb1 | -0.123 |
| igsf9a | 0.238 | srl | -0.123 |
| nid2a | 0.237 | neb | -0.123 |
| hoxb10a | 0.236 | pvalb2 | -0.122 |
| her7 | 0.236 | myl1 | -0.122 |
| adgrl2a | 0.232 | mylz3 | -0.122 |
| itm2cb | 0.232 | ldb3b | -0.121 |
| arl4aa | 0.231 | cav3 | -0.120 |
| fhdc2 | 0.229 | col1a1a | -0.120 |
| mespab | 0.228 | tnnt3a | -0.120 |
| hes6 | 0.228 | CABZ01078594.1 | -0.119 |
| rem1 | 0.225 | vwde | -0.118 |