"deltex 4, E3 ubiquitin ligase a"
ZFIN 
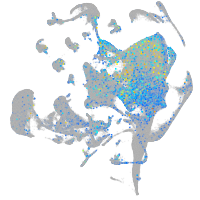
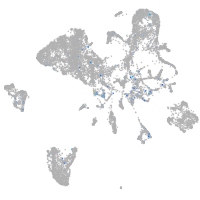
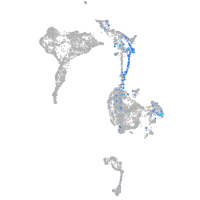
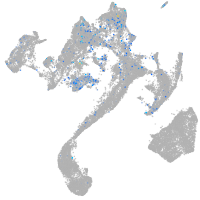
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU634008.1 | 0.238 | ptmaa | -0.105 |
| CU467822.1 | 0.231 | elavl3 | -0.096 |
| notch3 | 0.226 | cdx4 | -0.096 |
| CR848047.1 | 0.224 | stxbp1a | -0.085 |
| gpm6bb | 0.200 | hoxb10a | -0.084 |
| zgc:165461 | 0.184 | gng3 | -0.082 |
| chfr | 0.182 | hoxc3a | -0.082 |
| fabp7a | 0.182 | tuba8l3 | -0.081 |
| cd82a | 0.179 | si:ch211-222l21.1 | -0.081 |
| plp1a | 0.178 | hoxb9a | -0.080 |
| npas1 | 0.168 | fthl27 | -0.079 |
| si:ch73-215f7.1 | 0.158 | vamp2 | -0.079 |
| sox9b | 0.150 | sncb | -0.079 |
| fgfr2 | 0.148 | jagn1a | -0.078 |
| nrarpb | 0.148 | hoxb7a | -0.076 |
| msi1 | 0.146 | stx1b | -0.075 |
| ptprz1b | 0.145 | onecut1 | -0.075 |
| nfia | 0.144 | onecut2 | -0.075 |
| masp1 | 0.143 | lin28a | -0.074 |
| atp1b4 | 0.142 | LOC100537384 | -0.073 |
| ptprfb | 0.141 | apela | -0.072 |
| sox2 | 0.141 | scrt2 | -0.072 |
| si:ch1073-303k11.2 | 0.140 | elavl4 | -0.072 |
| notch1b | 0.140 | stmn1b | -0.072 |
| cspg5a | 0.139 | tmem59l | -0.071 |
| si:ch73-30b17.2 | 0.139 | snap25a | -0.071 |
| lrrn1 | 0.138 | hoxa9a | -0.071 |
| CU929451.2 | 0.137 | atp6v0cb | -0.070 |
| marcksl1a | 0.136 | calm1a | -0.070 |
| si:ch73-21g5.7 | 0.136 | nhlh2 | -0.067 |
| sox3 | 0.136 | nsg2 | -0.067 |
| BX664610.1 | 0.135 | cplx2 | -0.066 |
| metrnla | 0.135 | hoxd10a | -0.066 |
| tnc | 0.134 | hoxa10b | -0.066 |
| slc1a2b | 0.133 | npm1a | -0.066 |