dehydrogenase/reductase (SDR family) member 3b
ZFIN 
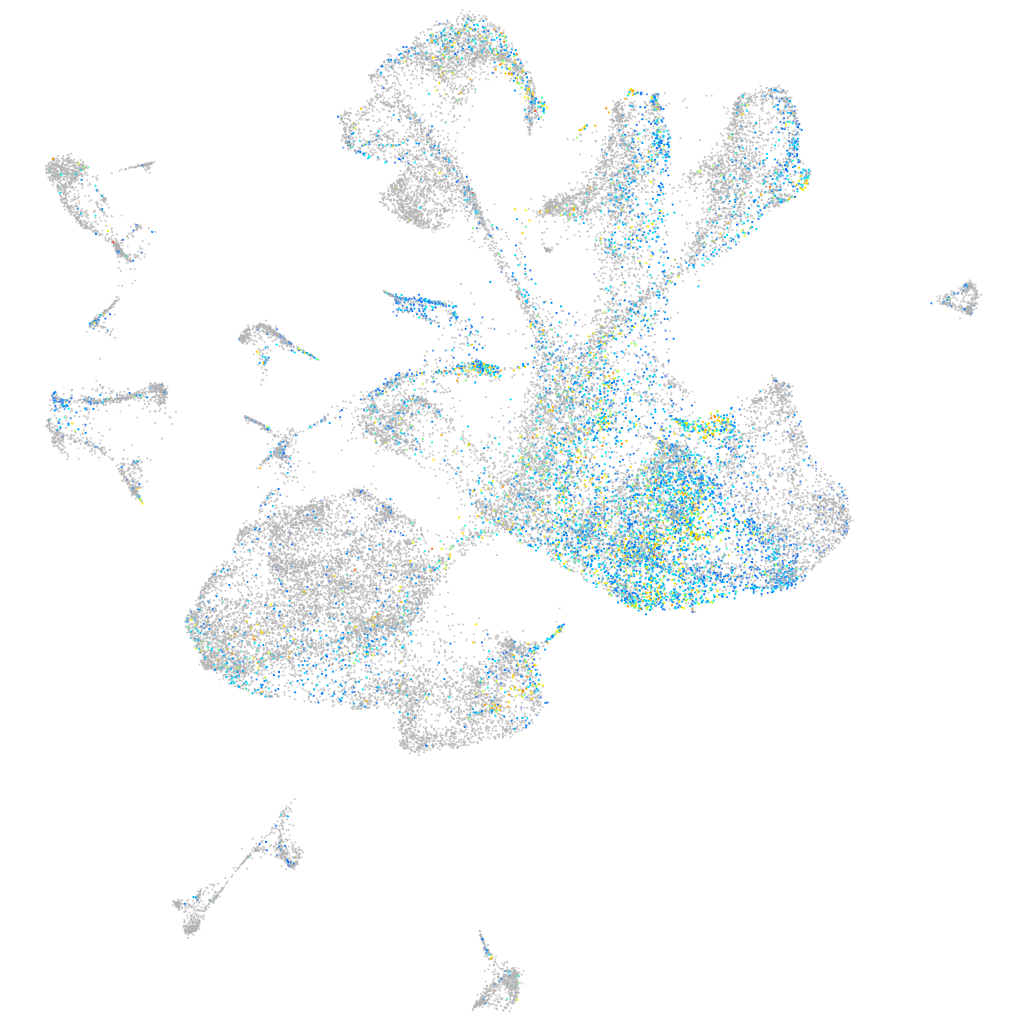
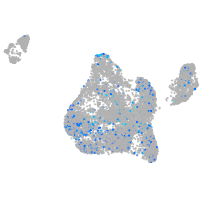
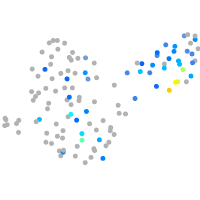
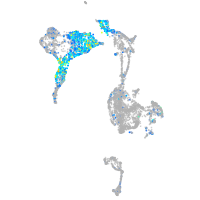
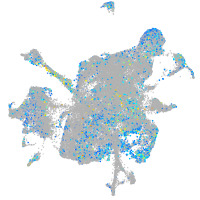
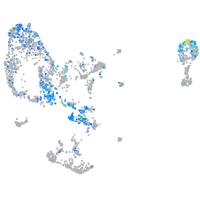
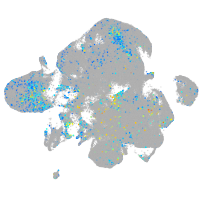
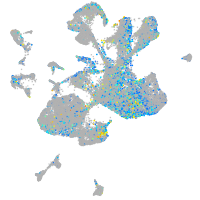
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxc1a | 0.390 | ckbb | -0.186 |
| slc1a3a | 0.331 | CU467822.1 | -0.160 |
| hoxb1b | 0.317 | pvalb1 | -0.149 |
| hoxb3a | 0.288 | gpm6ab | -0.143 |
| hoxc3a | 0.288 | pvalb2 | -0.138 |
| CABZ01075068.1 | 0.282 | ndrg2 | -0.123 |
| hoxb6b | 0.264 | gpm6aa | -0.116 |
| hoxb8a | 0.257 | CU634008.1 | -0.114 |
| hoxb5b | 0.254 | slc1a2b | -0.114 |
| cygb1 | 0.248 | COX3 | -0.112 |
| hoxb6a | 0.246 | mt-atp6 | -0.110 |
| dhrs3a | 0.245 | CR848047.1 | -0.110 |
| si:ch211-286o17.1 | 0.240 | celf2 | -0.109 |
| hoxa3a | 0.230 | nfia | -0.108 |
| raraa | 0.220 | gapdhs | -0.106 |
| cyp27c1 | 0.220 | nanos1 | -0.101 |
| nr2f6b | 0.217 | itm2ba | -0.100 |
| wnt11r | 0.211 | cadm4 | -0.100 |
| XLOC-003689 | 0.204 | fez1 | -0.099 |
| nr6a1b | 0.196 | actc1b | -0.097 |
| hmgb2a | 0.194 | acbd7 | -0.096 |
| hoxb8b | 0.186 | hoxd12a | -0.096 |
| CU571074.1 | 0.182 | elavl4 | -0.094 |
| tes | 0.179 | tnrc6c1 | -0.094 |
| fabp3 | 0.178 | rarga | -0.091 |
| her12 | 0.177 | hoxd10a | -0.090 |
| hmga1a | 0.176 | efhd1 | -0.089 |
| mapk12b | 0.176 | hbae3 | -0.089 |
| pax3a | 0.173 | glula | -0.088 |
| si:ch73-265d7.2 | 0.172 | nfil3-5 | -0.088 |
| meis1a | 0.168 | atp5if1b | -0.088 |
| nr2f5 | 0.167 | ptn | -0.087 |
| mir219-3 | 0.166 | ppap2d | -0.086 |
| h2afvb | 0.165 | cst3 | -0.086 |
| rgcc | 0.164 | sox4a | -0.086 |