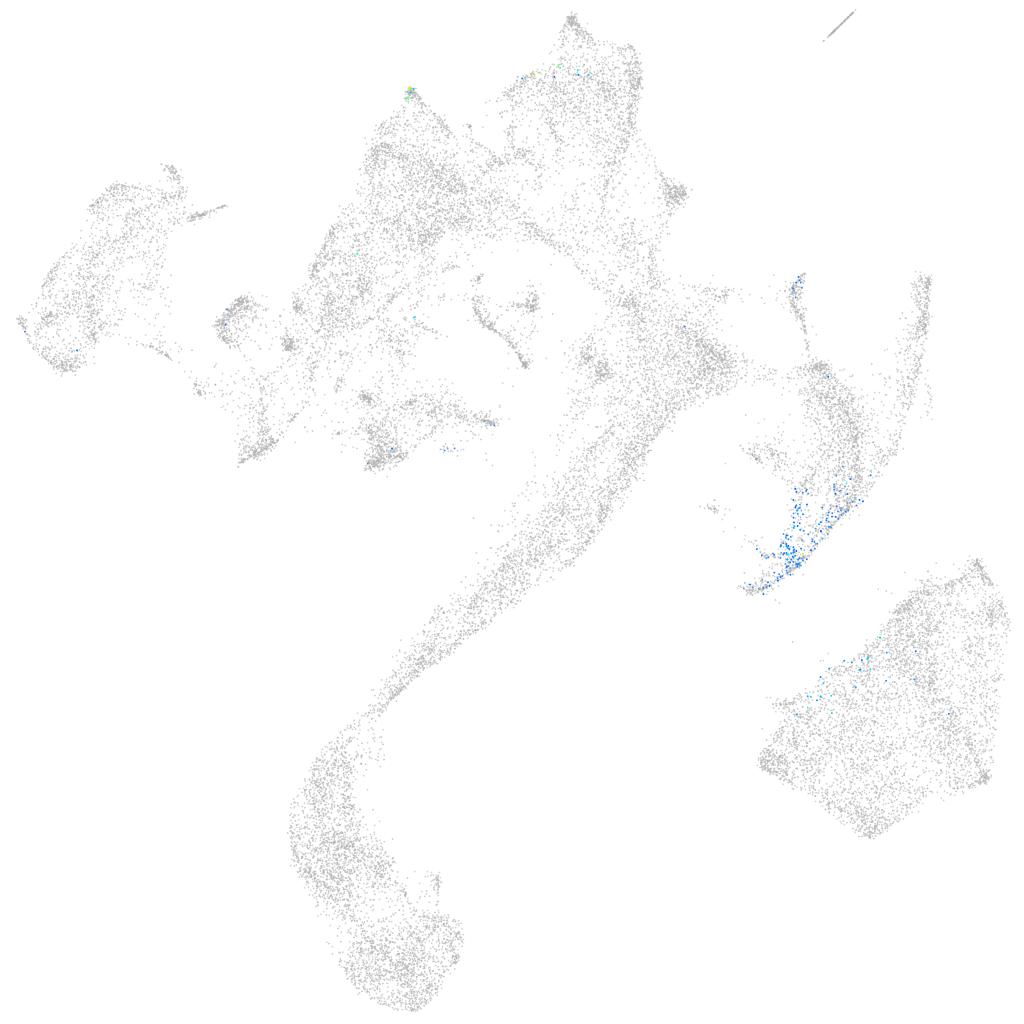
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 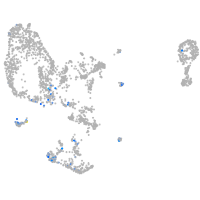 |
neural |
spinal cord / glia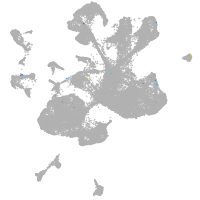 |
eye |
taste / olfactory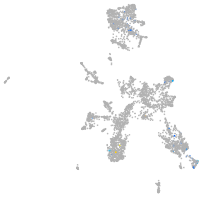 |
otic / lateral line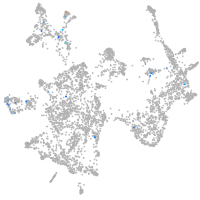 |
epidermis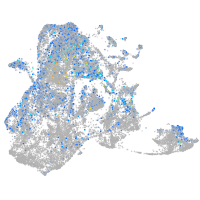 |
periderm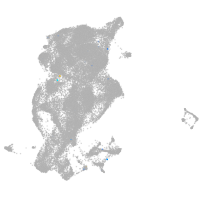 |
fin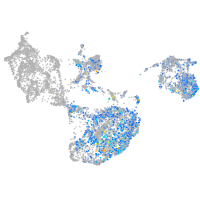 |
ionocytes / mucous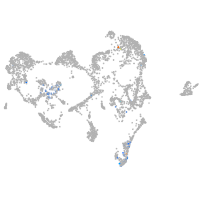 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fgf3 | 0.229 | ttn.2 | -0.061 |
| tagln3b | 0.217 | fabp3 | -0.060 |
| thbs2a | 0.201 | ttn.1 | -0.060 |
| gdf11 | 0.183 | actc1b | -0.055 |
| bmp2a | 0.182 | hsp90aa1.1 | -0.054 |
| hoxd12a | 0.173 | zgc:92429 | -0.052 |
| tll1 | 0.171 | aldoab | -0.050 |
| cyp26a1 | 0.171 | klhl41b | -0.050 |
| hoxa13b | 0.169 | tmem38a | -0.049 |
| itm2cb | 0.158 | fstl1a | -0.049 |
| pip5k1ca | 0.154 | pabpc4 | -0.049 |
| hoxd10a | 0.152 | six1b | -0.048 |
| zgc:162939 | 0.146 | mybphb | -0.048 |
| depdc7a | 0.144 | fxr2 | -0.048 |
| hoxb10a | 0.143 | emp2 | -0.048 |
| esrrga | 0.143 | zgc:92518 | -0.048 |
| hoxa11a | 0.139 | desma | -0.047 |
| hoxa10b | 0.135 | eno1a | -0.047 |
| zgc:100951 | 0.135 | atp2a1 | -0.047 |
| fgf8a | 0.134 | acta1a | -0.046 |
| b3gnt7l | 0.133 | tpma | -0.046 |
| hes6 | 0.130 | actc1a | -0.046 |
| vox | 0.129 | cpn1 | -0.046 |
| enc3 | 0.129 | tnnc2 | -0.046 |
| rdh8a | 0.127 | rbm24a | -0.046 |
| greb1 | 0.125 | tnni2b.1 | -0.045 |
| her7 | 0.125 | gatm | -0.045 |
| her12 | 0.124 | ckma | -0.045 |
| hoxc11a | 0.122 | ak1 | -0.045 |
| col5a3a | 0.122 | rbfox1l | -0.045 |
| sall4 | 0.122 | cox17 | -0.045 |
| tbxtb | 0.118 | nr2f5 | -0.045 |
| hoxd11a | 0.118 | CABZ01078594.1 | -0.044 |
| si:ch211-255i20.3 | 0.117 | meis1a | -0.044 |
| vent | 0.116 | nexn | -0.044 |