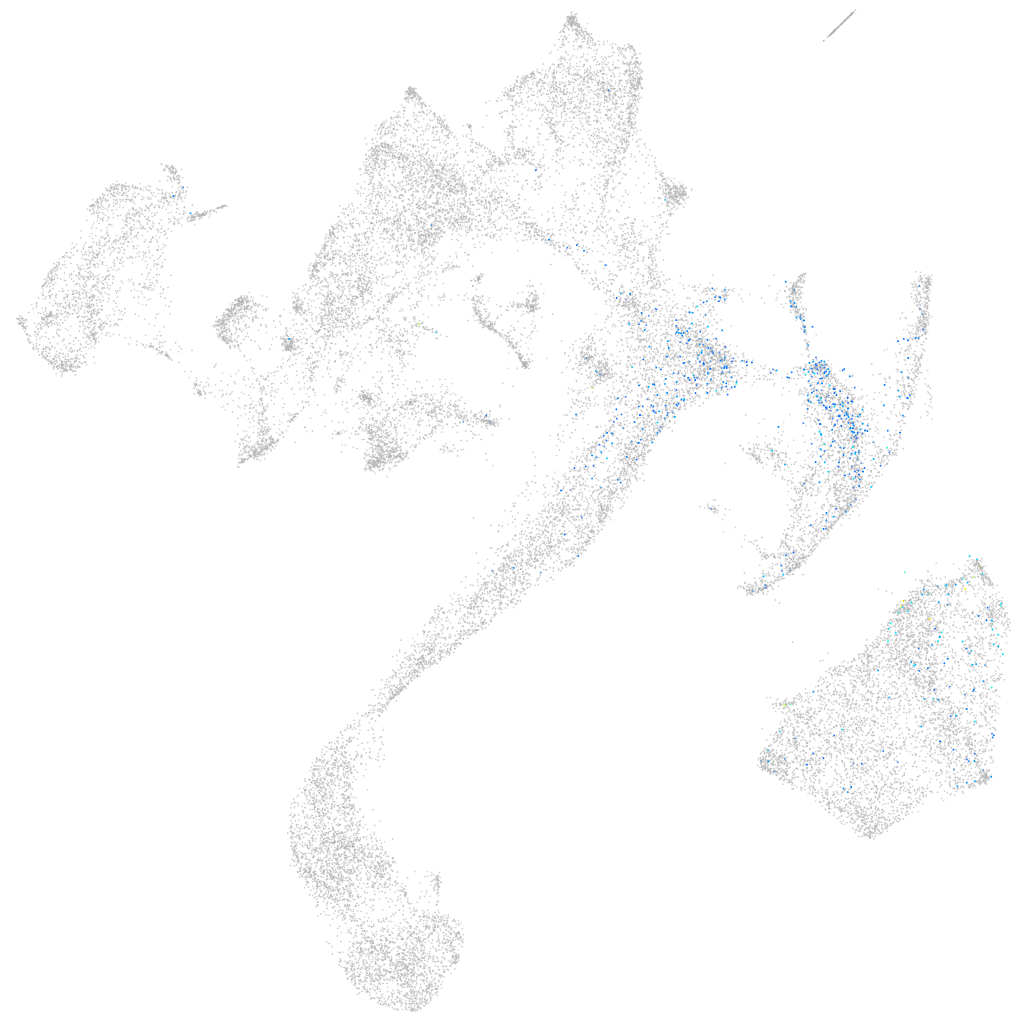
zinc finger-like gene 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| znfl1k | 0.163 | actc1b | -0.095 |
| tbx6 | 0.155 | fabp3 | -0.091 |
| ripply1 | 0.149 | bhmt | -0.090 |
| fn1b | 0.149 | ckma | -0.084 |
| apoc1 | 0.147 | atp2a1 | -0.084 |
| draxin | 0.146 | ckmb | -0.083 |
| meox1 | 0.139 | ak1 | -0.081 |
| ripply2 | 0.139 | mylpfa | -0.080 |
| myf5 | 0.138 | aldoab | -0.080 |
| pcdh8 | 0.138 | pabpc4 | -0.079 |
| BX001014.2 | 0.133 | tmem38a | -0.079 |
| gnaia | 0.131 | tnnc2 | -0.077 |
| tjp2b | 0.130 | acta1b | -0.077 |
| her11 | 0.125 | pmp22a | -0.076 |
| nid2a | 0.123 | sparc | -0.076 |
| tcf15 | 0.119 | gapdh | -0.076 |
| her1 | 0.119 | tpi1b | -0.076 |
| apoeb | 0.119 | eno1a | -0.075 |
| dlc | 0.114 | neb | -0.075 |
| si:ch73-281n10.2 | 0.113 | si:dkey-16p21.8 | -0.074 |
| kazald2 | 0.113 | pvalb1 | -0.074 |
| BX005254.3 | 0.113 | srl | -0.073 |
| AL928824.1 | 0.113 | pvalb2 | -0.073 |
| npm1a | 0.110 | col1a2 | -0.073 |
| nhp2 | 0.109 | gamt | -0.073 |
| samd10b | 0.109 | ndrg2 | -0.072 |
| asph | 0.108 | eno3 | -0.072 |
| lmo4b | 0.106 | mylz3 | -0.072 |
| BX927258.1 | 0.106 | ldb3b | -0.072 |
| efnb2b | 0.106 | desma | -0.071 |
| XLOC-003984 | 0.106 | tpma | -0.071 |
| grin2da | 0.105 | myl1 | -0.070 |
| mespab | 0.105 | si:ch73-367p23.2 | -0.070 |
| tuba8l2 | 0.105 | si:ch211-140m22.7 | -0.070 |
| nop58 | 0.104 | ldb3a | -0.069 |