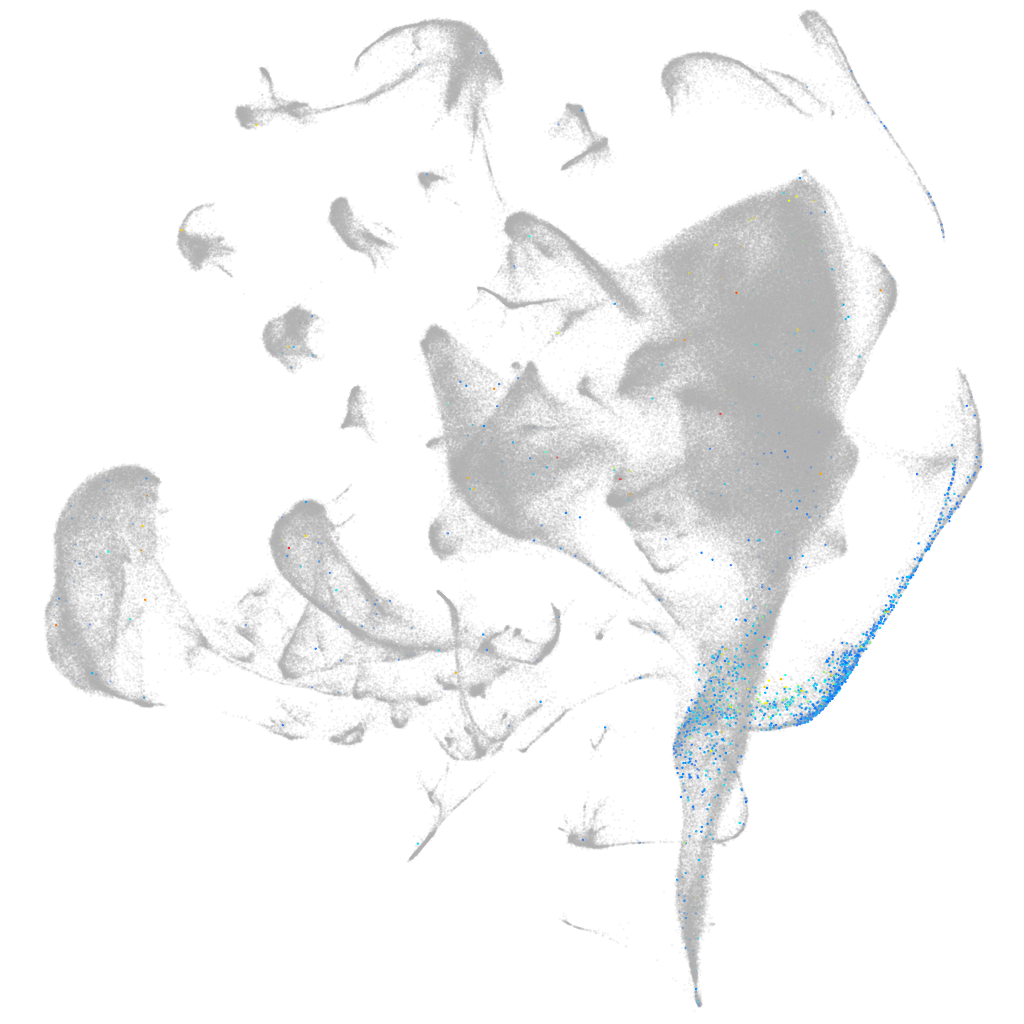
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 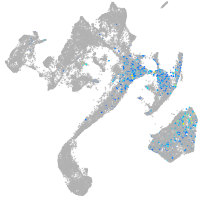 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 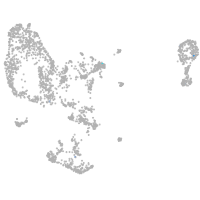 |
neural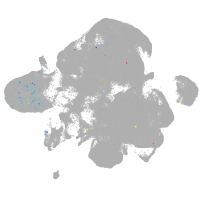 |
spinal cord / glia |
eye |
taste / olfactory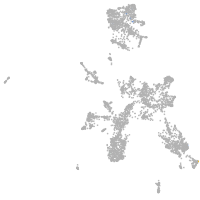 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tbx6 | 0.203 | fabp3 | -0.046 |
| znfl1k | 0.198 | tuba1c | -0.041 |
| myf5 | 0.195 | marcksl1a | -0.039 |
| tcf15 | 0.173 | pvalb1 | -0.038 |
| XLOC-042229 | 0.170 | pvalb2 | -0.037 |
| meox1 | 0.159 | gapdhs | -0.035 |
| LOC110438394 | 0.156 | gpm6aa | -0.035 |
| XLOC-040516 | 0.148 | prdx2 | -0.035 |
| hsp90aa1.1 | 0.145 | hbbe1.3 | -0.033 |
| XLOC-009784 | 0.141 | krt4 | -0.033 |
| XLOC-003984 | 0.140 | ckbb | -0.032 |
| XLOC-004280 | 0.139 | nova2 | -0.032 |
| ripply2 | 0.136 | rtn1a | -0.032 |
| BX001014.2 | 0.135 | wu:fb55g09 | -0.032 |
| unc45b | 0.132 | calm1a | -0.032 |
| ripply1 | 0.130 | ccni | -0.031 |
| zgc:92429 | 0.130 | elavl3 | -0.031 |
| BX276101.1 | 0.125 | hbae3 | -0.031 |
| dmrt2a | 0.123 | celf2 | -0.030 |
| mespaa | 0.123 | hbae1.1 | -0.030 |
| im:7138239 | 0.122 | rnasekb | -0.030 |
| BX927258.1 | 0.121 | stmn1b | -0.030 |
| mespab | 0.120 | cyt1 | -0.029 |
| her1 | 0.120 | fabp7a | -0.029 |
| mespba | 0.119 | foxp4 | -0.029 |
| apoc1 | 0.116 | eif4a1b | -0.028 |
| fgf17 | 0.112 | mdkb | -0.028 |
| hspb1 | 0.112 | tmsb | -0.028 |
| msgn1 | 0.112 | her15.1 | -0.028 |
| znfl1h | 0.111 | cyt1l | -0.027 |
| fn1b | 0.110 | gpm6ab | -0.027 |
| LOC103910903 | 0.110 | icn | -0.027 |
| znfl1 | 0.109 | cotl1 | -0.026 |
| kazald2 | 0.108 | mylpfa | -0.026 |
| myod1 | 0.107 | tuba1a | -0.026 |