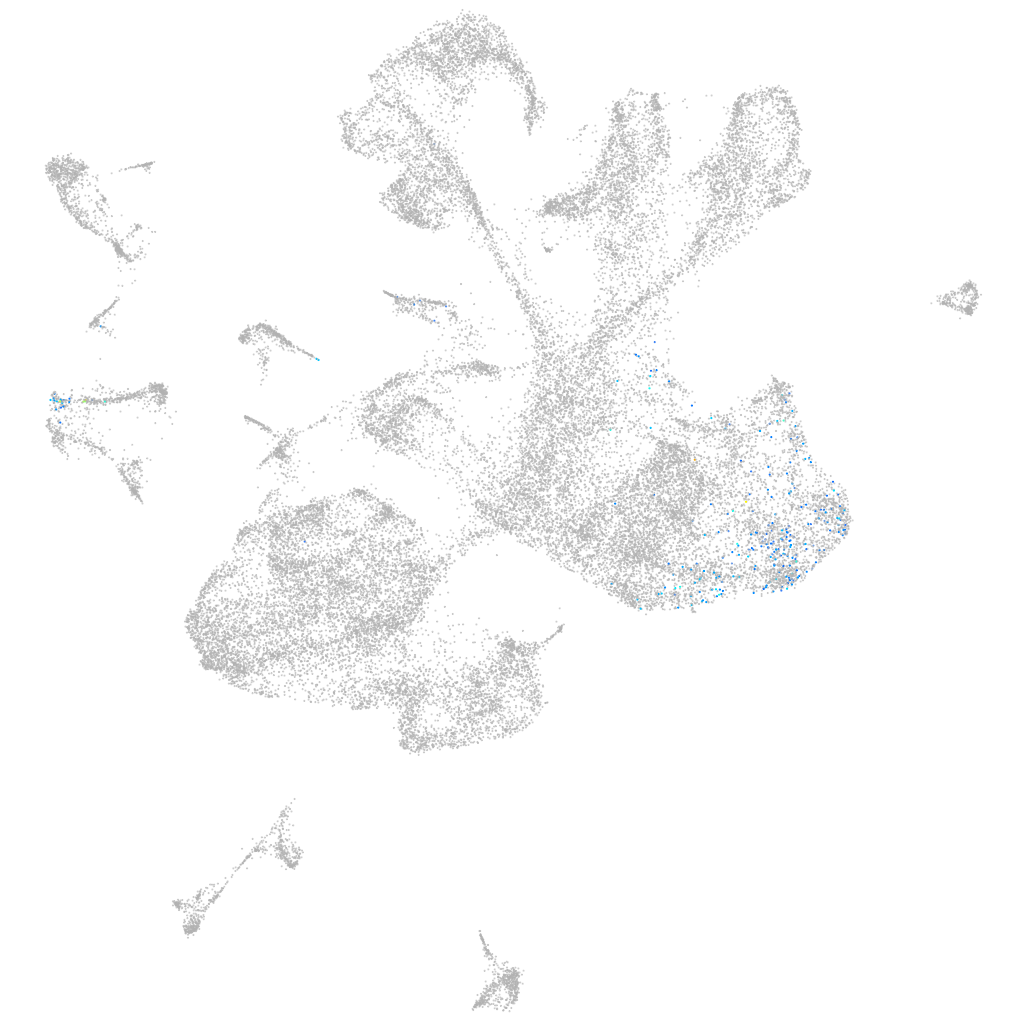
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 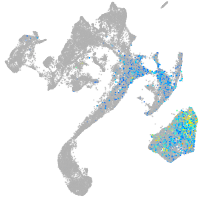 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 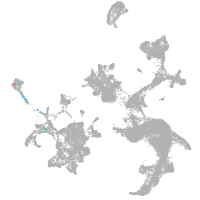 |
pronephros 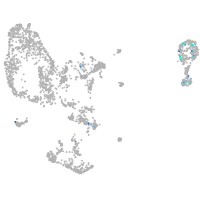 |
neural |
spinal cord / glia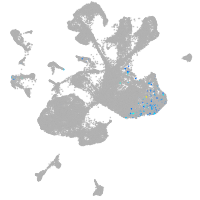 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdx4 | 0.186 | gpm6aa | -0.080 |
| wnt11r | 0.146 | ckbb | -0.071 |
| BX001014.2 | 0.137 | fabp7a | -0.061 |
| gdf6b | 0.134 | rtn1a | -0.059 |
| XLOC-042222 | 0.128 | pvalb1 | -0.059 |
| apela | 0.128 | CU467822.1 | -0.059 |
| hspb1 | 0.124 | pvalb2 | -0.058 |
| sall4 | 0.121 | tmsb4x | -0.055 |
| nradd | 0.113 | hbbe1.3 | -0.054 |
| pcolce2b | 0.112 | tmsb | -0.054 |
| XLOC-001964 | 0.110 | COX3 | -0.053 |
| cpdb | 0.109 | gpm6ab | -0.053 |
| msx1b | 0.107 | zgc:165461 | -0.052 |
| BX927258.1 | 0.107 | marcksl1b | -0.051 |
| oc90 | 0.106 | CU634008.1 | -0.051 |
| ldlrad2 | 0.103 | CR383676.1 | -0.050 |
| mllt3 | 0.101 | elavl3 | -0.049 |
| BX005254.3 | 0.099 | actc1b | -0.049 |
| npm1a | 0.099 | gpm6bb | -0.047 |
| angptl2b | 0.098 | myt1b | -0.047 |
| sfrp2 | 0.095 | vim | -0.047 |
| zic2b | 0.094 | hbae3 | -0.046 |
| si:ch211-170d8.2 | 0.093 | gapdhs | -0.046 |
| wu:fb97g03 | 0.093 | nova2 | -0.046 |
| edn1 | 0.091 | gnao1a | -0.045 |
| irx1b | 0.090 | stmn1b | -0.045 |
| irx3b | 0.090 | si:dkeyp-75h12.5 | -0.044 |
| dkc1 | 0.089 | dpysl2b | -0.044 |
| thbs3b | 0.088 | atp1a1b | -0.044 |
| notum1a | 0.087 | rnasekb | -0.043 |
| si:dkey-102m7.3 | 0.087 | slc1a2b | -0.043 |
| XLOC-012749 | 0.086 | mylpfa | -0.043 |
| asb11 | 0.086 | si:ch211-251b21.1 | -0.042 |
| arid3b | 0.085 | si:ch73-21g5.7 | -0.041 |
| znfl1k | 0.085 | fez1 | -0.040 |