ubiquitin specific peptidase 43a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm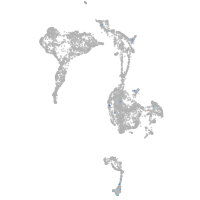 |
muscle 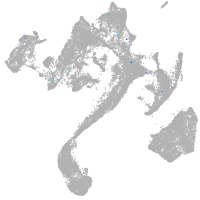 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 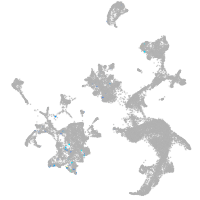 |
pronephros 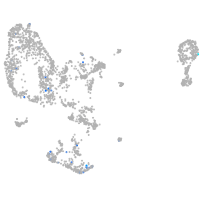 |
neural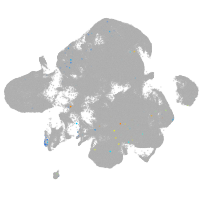 |
spinal cord / glia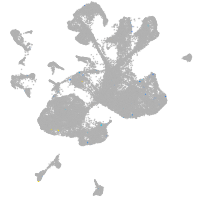 |
eye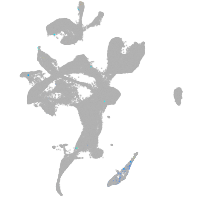 |
taste / olfactory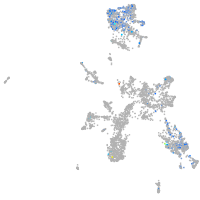 |
otic / lateral line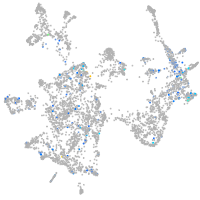 |
epidermis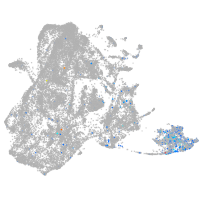 |
periderm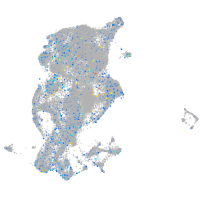 |
fin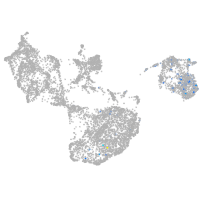 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lim2.4 | 0.134 | ptmab | -0.069 |
| cryba2a | 0.131 | hmgb2a | -0.052 |
| gja8b | 0.131 | hmgn6 | -0.050 |
| mipa | 0.131 | h2afva | -0.045 |
| crybb1 | 0.129 | hmga1a | -0.045 |
| mipb | 0.128 | si:ch73-1a9.3 | -0.042 |
| b3gnt5a | 0.127 | hnrnpaba | -0.041 |
| cryba4 | 0.127 | hmgb2b | -0.041 |
| gja3 | 0.126 | hmgn2 | -0.039 |
| crybb1l1 | 0.126 | h3f3a | -0.038 |
| cryba1b | 0.125 | si:ch211-222l21.1 | -0.037 |
| aifm5 | 0.125 | hnrnpa0b | -0.037 |
| crygn2 | 0.124 | h2afvb | -0.037 |
| cryba2b | 0.123 | khdrbs1a | -0.036 |
| cx23 | 0.122 | fabp7a | -0.036 |
| cryba1l1 | 0.122 | hnrnpabb | -0.035 |
| crybb1l2 | 0.121 | hmgb1b | -0.035 |
| prx | 0.120 | hmgb1a | -0.034 |
| htr2aa | 0.117 | cirbpb | -0.034 |
| lim2.3 | 0.117 | fabp3 | -0.034 |
| BFSP1 | 0.116 | si:ch211-137a8.4 | -0.033 |
| lim2.1 | 0.116 | h3f3d | -0.030 |
| cps1 | 0.115 | rbm8a | -0.030 |
| crygmx | 0.114 | tuba1a | -0.030 |
| prox2 | 0.114 | tubb2b | -0.029 |
| otc | 0.113 | marcksb | -0.029 |
| cryba1l2 | 0.111 | actb2 | -0.029 |
| gja8a | 0.109 | erh | -0.028 |
| LOC101885188 | 0.106 | chd4a | -0.028 |
| rbm24a | 0.103 | ddx39ab | -0.028 |
| si:ch211-214j24.14 | 0.103 | cx43.4 | -0.027 |
| gpx9 | 0.103 | sumo3a | -0.027 |
| tubb6 | 0.102 | alyref | -0.027 |
| BX465225.1 | 0.102 | rbbp4 | -0.027 |
| xirp1 | 0.100 | snrpf | -0.027 |




