T-box transcription factor 20
ZFIN 
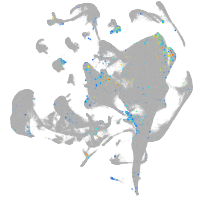
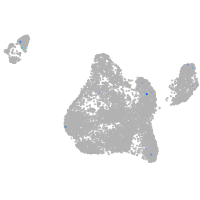
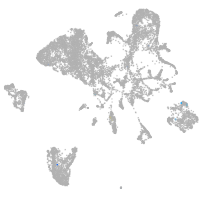
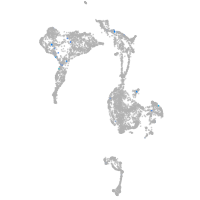
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| phox2bb | 0.571 | mdka | -0.185 |
| phox2a | 0.569 | id1 | -0.182 |
| lmo4b | 0.445 | XLOC-003690 | -0.180 |
| isl1 | 0.404 | her15.1 | -0.161 |
| CR812832.1 | 0.376 | cldn5a | -0.153 |
| prph | 0.366 | notch3 | -0.150 |
| slit3 | 0.362 | sox3 | -0.147 |
| tbx2b | 0.355 | XLOC-003689 | -0.131 |
| map1b | 0.330 | cd82a | -0.127 |
| tbx3a | 0.301 | vamp3 | -0.127 |
| gap43 | 0.297 | sdc4 | -0.126 |
| mab21l2 | 0.294 | sox19a | -0.126 |
| onecut1 | 0.293 | hoxb9a | -0.125 |
| nkx6.1 | 0.287 | gfap | -0.124 |
| chodl | 0.273 | btg2 | -0.124 |
| nefmb | 0.269 | CU467822.1 | -0.123 |
| cbfb | 0.268 | fabp7a | -0.123 |
| shox | 0.266 | COX7A2 (1 of many) | -0.123 |
| isl2a | 0.266 | atp1a1b | -0.122 |
| tubb5 | 0.265 | msi1 | -0.122 |
| meis2b | 0.257 | sox2 | -0.120 |
| slc18a3a | 0.256 | her4.1 | -0.118 |
| rap1gap | 0.249 | her12 | -0.118 |
| inab | 0.249 | sparc | -0.117 |
| stmn1b | 0.248 | gpm6bb | -0.116 |
| stmn4l | 0.247 | sb:cb81 | -0.115 |
| rtn1b | 0.247 | si:ch211-286b5.5 | -0.113 |
| stmn2a | 0.246 | hmgb2a | -0.113 |
| mmp17a | 0.245 | gas1b | -0.111 |
| vip | 0.241 | XLOC-003692 | -0.111 |
| tppp2 | 0.237 | zgc:165461 | -0.109 |
| id4 | 0.235 | tuba8l | -0.109 |
| mllt11 | 0.234 | her4.2 | -0.108 |
| neflb | 0.233 | hoxc3a | -0.108 |
| nrp2a | 0.233 | tspan7 | -0.107 |