"TBC1 domain family, member 17"
ZFIN 
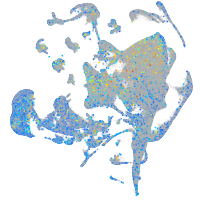
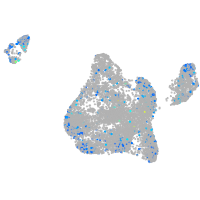
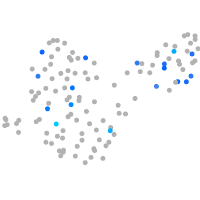
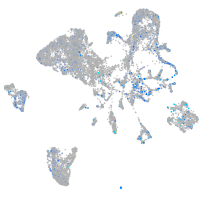
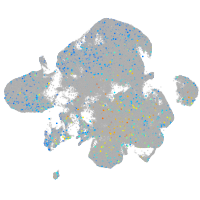
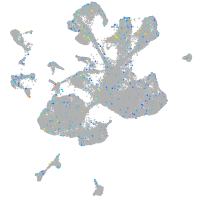
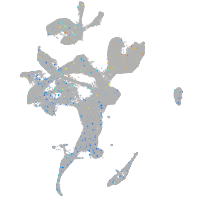
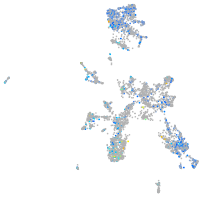
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX890543.1 | 0.169 | rplp1 | -0.132 |
| si:dkey-192d15.2 | 0.159 | si:ch211-222l21.1 | -0.132 |
| atp6ap1b | 0.159 | si:dkey-151g10.6 | -0.124 |
| ceacam1 | 0.157 | rplp2l | -0.120 |
| soul2 | 0.157 | stmn1a | -0.113 |
| cox5b2 | 0.156 | hmgb2b | -0.109 |
| ca15a | 0.155 | ptmaa | -0.107 |
| ca2 | 0.155 | ptmab | -0.104 |
| rnf128a | 0.155 | rps29 | -0.103 |
| atp6v1h | 0.152 | hmgb1b | -0.102 |
| eps8l1b | 0.152 | rps27.1 | -0.102 |
| slc4a1b | 0.151 | setb | -0.101 |
| atp6v0ca | 0.151 | si:ch211-288g17.3 | -0.098 |
| atp6v0b | 0.151 | rpl37 | -0.097 |
| rnaseka | 0.150 | rps21 | -0.095 |
| abca12 | 0.149 | rpl31 | -0.095 |
| clcn2b | 0.149 | snrpd2 | -0.094 |
| atp6v1aa | 0.146 | snrpf | -0.093 |
| LOC100537613 | 0.146 | rps14 | -0.091 |
| zgc:193726 | 0.145 | rcc2 | -0.090 |
| mylk5 | 0.143 | seta | -0.090 |
| enosf1 | 0.141 | marcksb | -0.090 |
| gpha2 | 0.141 | tmsb4x | -0.090 |
| rhcgb | 0.141 | apex1 | -0.089 |
| gb:bc139872 | 0.140 | hmgn2 | -0.089 |
| atp6v0a1a | 0.140 | lmnb2 | -0.089 |
| zgc:56622 | 0.140 | eif4a1a | -0.087 |
| ISCU (1 of many) | 0.139 | zc4h2 | -0.087 |
| atp6v1g1 | 0.138 | rpl38 | -0.087 |
| atp6v0d1 | 0.138 | rpl26 | -0.086 |
| si:dkeyp-72e1.7 | 0.137 | dut | -0.086 |
| atp6v1e1b | 0.136 | snu13b | -0.086 |
| atp6v1c1b | 0.136 | ran | -0.085 |
| ly6m2 | 0.135 | tubb2b | -0.085 |
| epdl1 | 0.134 | si:ch211-39i22.1 | -0.085 |