"superoxide dismutase 1, soluble"
ZFIN 
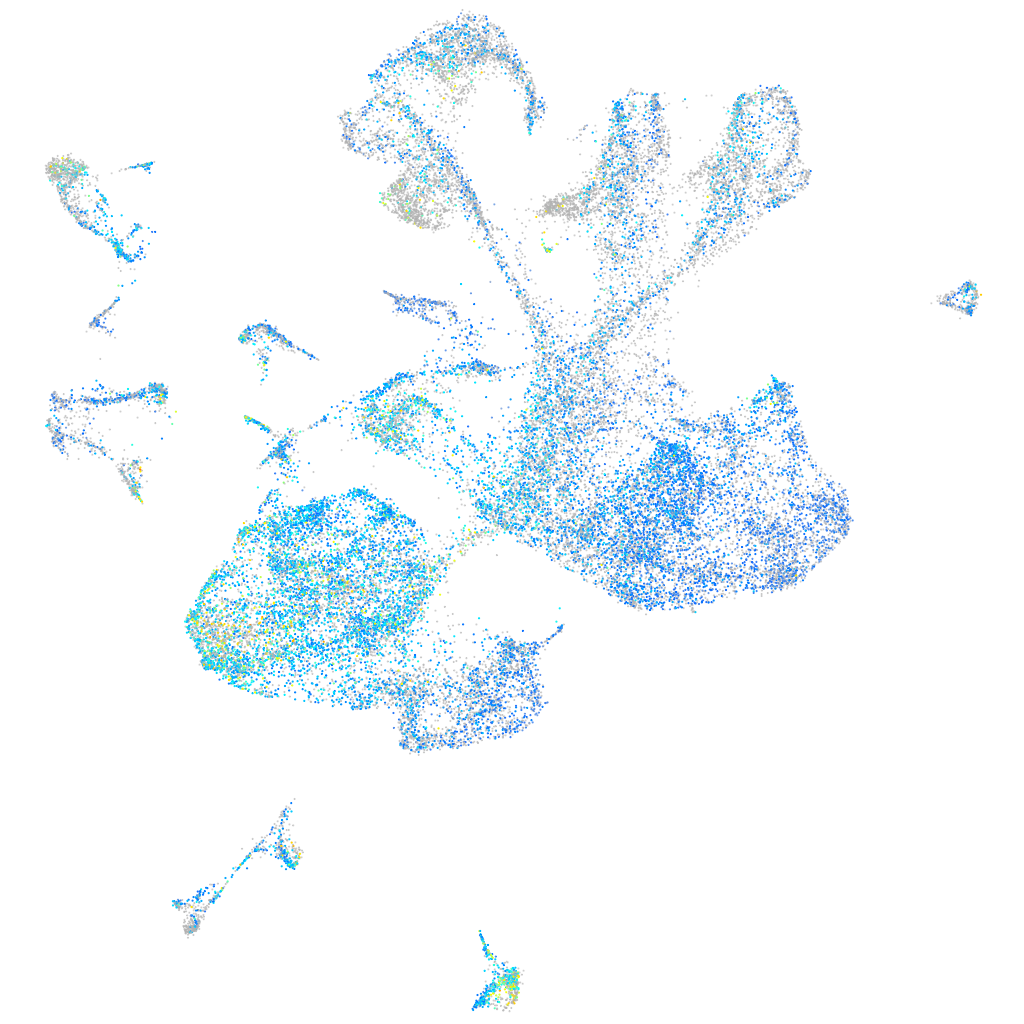
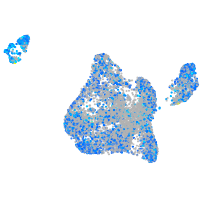
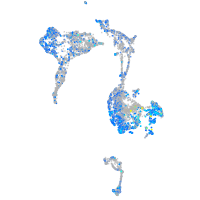
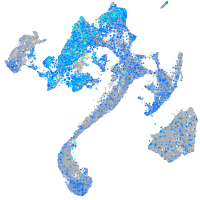
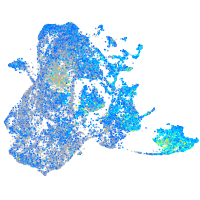
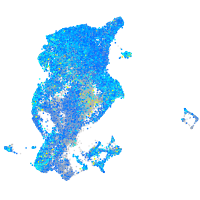
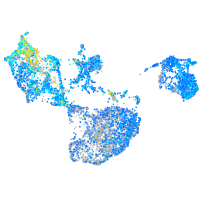
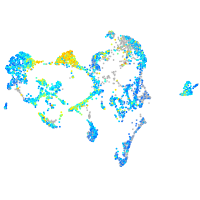
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fabp7a | 0.318 | elavl3 | -0.217 |
| CU467822.1 | 0.292 | myt1b | -0.170 |
| CR848047.1 | 0.260 | onecut1 | -0.160 |
| ppap2d | 0.237 | lin28a | -0.158 |
| atp1a1b | 0.232 | hoxb9a | -0.153 |
| atp1b4 | 0.230 | pik3r3b | -0.152 |
| slc1a2b | 0.230 | si:ch211-222l21.1 | -0.151 |
| zgc:165461 | 0.224 | onecut2 | -0.148 |
| gpm6bb | 0.221 | zgc:100920 | -0.146 |
| glula | 0.216 | si:ch73-386h18.1 | -0.144 |
| psat1 | 0.209 | myt1a | -0.144 |
| nfia | 0.207 | hoxc3a | -0.143 |
| mdka | 0.201 | gng2 | -0.141 |
| efhd1 | 0.201 | zc4h2 | -0.140 |
| slc3a2a | 0.200 | LOC100537384 | -0.139 |
| si:ch211-66e2.5 | 0.198 | syt2a | -0.139 |
| mdkb | 0.194 | zfhx3 | -0.138 |
| cspg5b | 0.192 | jagn1a | -0.136 |
| slc1a3b | 0.190 | stxbp1a | -0.134 |
| cd82a | 0.189 | ptmab | -0.131 |
| GCA | 0.189 | tubb5 | -0.130 |
| cdo1 | 0.188 | h3f3d | -0.129 |
| id1 | 0.186 | CABZ01075068.1 | -0.129 |
| cd63 | 0.186 | scrt2 | -0.128 |
| hepacama | 0.182 | stx1b | -0.126 |
| prdx2 | 0.180 | thsd7aa | -0.126 |
| si:ch1073-303k11.2 | 0.179 | hoxb3a | -0.125 |
| itm2ba | 0.179 | elavl4 | -0.125 |
| ddt | 0.179 | nsg2 | -0.125 |
| cx43 | 0.179 | meis2b | -0.123 |
| gnai2a | 0.177 | rbfox1 | -0.123 |
| msi1 | 0.177 | tmeff1b | -0.123 |
| si:dkey-7j14.6 | 0.176 | epb41a | -0.121 |
| mif | 0.174 | NC-002333.4 | -0.120 |
| slc6a11b | 0.174 | snap25a | -0.120 |